Ann Clin Microbiol.
2019 Dec;22(4):105-109. 10.5145/ACM.2019.22.4.105.
Cellulitis Caused by a Novel Cupriavidus Species Strain J1218 Identified by Whole Genome Sequencing
- Affiliations
-
- 1Department of Laboratory Medicine, Chosun University Hospital, Gwangju, Korea. md.seraph@gmail.com
- 2Department of Laboratory Medicine, Korea University College of Medicine, Seoul, Korea.
- KMID: 2466397
- DOI: http://doi.org/10.5145/ACM.2019.22.4.105
Abstract
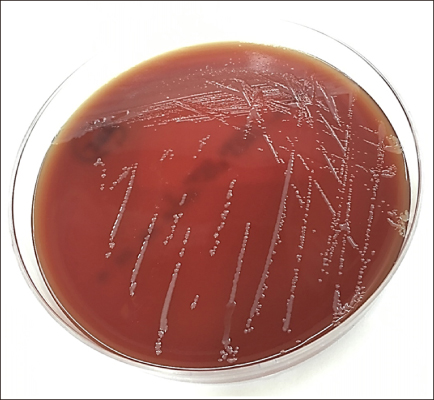
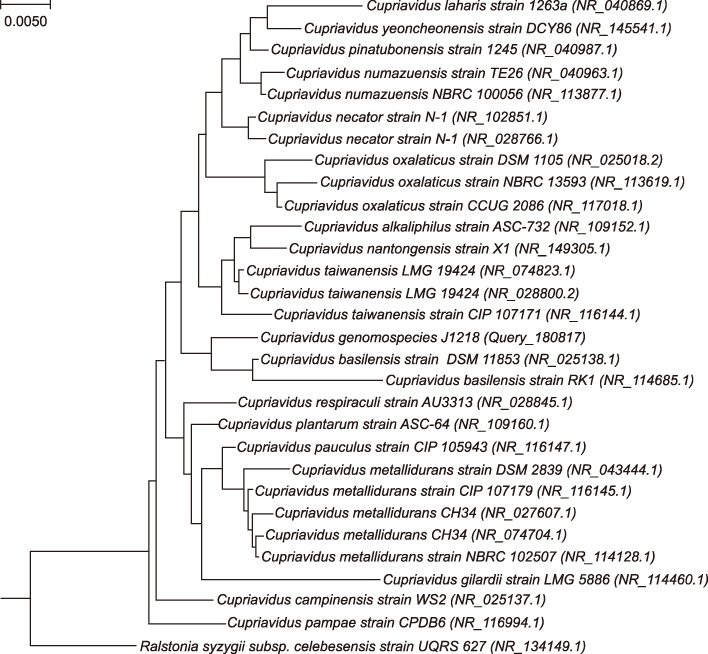
- We report a case of cellulitis caused by a novel Cupriavidus species identified using whole-genome sequence analysis. Subcutaneous tissue biopsies from the left lower leg of a 67-year-old man who suffered from cellulitis were cultured. Round, convex, gray and non-hemolytic colonies were recovered after 72-h incubation. 16S rRNA sequence analysis showed 98.6% similarity with Cupriavidus basilensis DSM 11853(T) in the NCBI database and 99.9% similarity with C. basilensis KF708 in the EzBioCloud database. Genomic analysis using the MiSeq platform (Illumina, USA) and the TrueBac ID database (ChunLab, Korea) revealed that the average nucleotide identity (ANI) of this strain with C. basilensis DSM 11853(T) was 87.6%. The patient was treated with oral cefditoren pivoxil for 9 weeks. This study is the first to report cellulitis caused by Cupriavidus species strain J1218.
Keyword
Figure
Reference
-
1. Duggal S, Gur R, Nayar R, Rongpharpi SR, Jain D, Gupta RK. Cupriavidus pauculus (Ralstonia paucula) concomitant meningitis and septicemia in a neonate: first case report from India. Indian J Med Microbiol. 2013; 31:405–409.2. D'Inzeo T, Santangelo R, Fiori B, De Angelis G, Conte V, Giaquinto A, et al. Catheter-related bacteremia by Cupriavidus metallidurans. Diagn Microbiol Infect Dis. 2015; 81:9–12.3. Almasy E, Szederjesi J, Rad P, Georgescu A. A fatal case of community acquired Cupriavidus pauculus pneumonia. J Crit Care Med (Targu Mures). 2016; 2:201–204.4. Kalka-Moll WM, LiPuma JJ, Accurso FJ, Plum G, van Koningsbruggen S, Vandamme P. Airway infection with a novel Cupriavidus species in persons with cystic fibrosis. J Clin Microbiol. 2009; 47:3026–3028.5. Karafin M, Romagnoli M, Fink DL, Howard T, Rau R, Milstone AM, et al. Fatal infection caused by Cupriavidus gilardii in a child with aplastic anemia. J Clin Microbiol. 2010; 48:1005–1007.6. Yahya R, Alyousef W, Omara A, Alamoudi S, Alshami A, Abdalhamid B. First case of pneumonia caused by Cupriavidus pauculus in an infant in the Gulf Cooperation Council. J Infect Dev Ctries. 2017; 11:196–198.7. Tena D, Losa C, Medina MJ, Sáez-Nieto JA. Muscular abscess caused by Cupriavidus gilardii in a renal transplant recipient. Diagn Microbiol Infect Dis. 2014; 79:108–110.8. Rosselló-Mora R, Amann R. The species concept for prokaryotes. FEMS Microbiol Rev. 2001; 25:39–67.9. Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, da Costa MS, et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol. 2018; 68:461–466.10. Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009; 106:19126–19131.11. Roach DJ, Burton JN, Lee C, Stackhouse B, Butler-Wu SM, Cookson BT, et al. A year of infection in the intensive care unit: prospective whole genome sequencing of bacterial clinical isolates reveals cryptic transmissions and novel microbiota. PLoS Genet. 2015; 11:e1005413.12. CLSI. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing. CLSI document MM18. Wayne, PA: Clinical and Laboratory Standards Institute;2018.13. Moriuchi R, Dohra H, Kanesaki Y, Ogawa N. Complete genome sequence of 3-chlorobenzoate-degrading bacterium Cupriavidus necator NH9 and reclassification of the strains of the genera Cupriavidus and Ralstonia based on phylogenetic and whole-genome sequence analyses. Front Microbiol. 2019; 10:133.14. CLSI. Performance standards for antimicrobial susceptibility testing. CLSI document M100. Wayne, PA: Clinical and Laboratory Standards Institute;2018.15. Zhang Z, Deng W, Wang S, Xu L, Yan L, Liao P. First case report of infection caused by Cupriavidus gilardii in a nonimmunocompromised Chinese patient. IDCases. 2017; 10:127–129.16. Kobayashi T, Nakamura I, Fujita H, Tsukimori A, Sato A, Fukushima S, et al. First case report of infection due to Cupriavidus gilardii in a patient without immunodeficiency: a case report. BMC Infect Dis. 2016; 16:493.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A Novel Species of the Genus Arsenicicoccus Isolated From Human Blood Using Whole-Genome Sequencing
- Whole genome sequence analyses of thermotolerant Bacillus sp. isolates from food
- A Case of Whole Genome Analysis of SARSCoV-2 Using Oxford Nanopore MinION System
- Genome-based identification of strain KCOM 1265 isolated from subgingival plaque at the species level
- Draft Genome Sequence of the White-Rot Fungus Schizophyllum commune IUM1114-SS01