Int J Stem Cells.
2019 Nov;12(3):430-439. 10.15283/ijsc19067.
Reprogramming of Cancer Cells into Induced Pluripotent Stem Cells Questioned
- Affiliations
-
- 1Department of Stem Cell Biology, Konkuk University School of Medicine, Seoul, Korea. knko@kku.ac.kr
- 2Center for Stem Cell Research, Institute of Advanced Biomedical Science, Konkuk University, Seoul, Korea.
- 3Department of Medicine, College of Medicine, Chung-Ang University, Seoul, Korea.
- 4Research Institute of Medical Science, Konkuk University, Seoul, Korea.
- KMID: 2465883
- DOI: http://doi.org/10.15283/ijsc19067
Abstract
- BACKGROUND AND OBJECTIVES
Several recent studies have claimed that cancer cells can be reprogrammed into induced pluripotent stem cells (iPSCs). However, in most cases, cancer cells seem to be resistant to cellular reprogramming. Furthermore, the underlying mechanisms of limited reprogramming in cancer cells are largely unknown. Here, we identified the candidate barrier genes and their target genes at the early stage of reprogramming for investigating cancer reprogramming.
METHODS
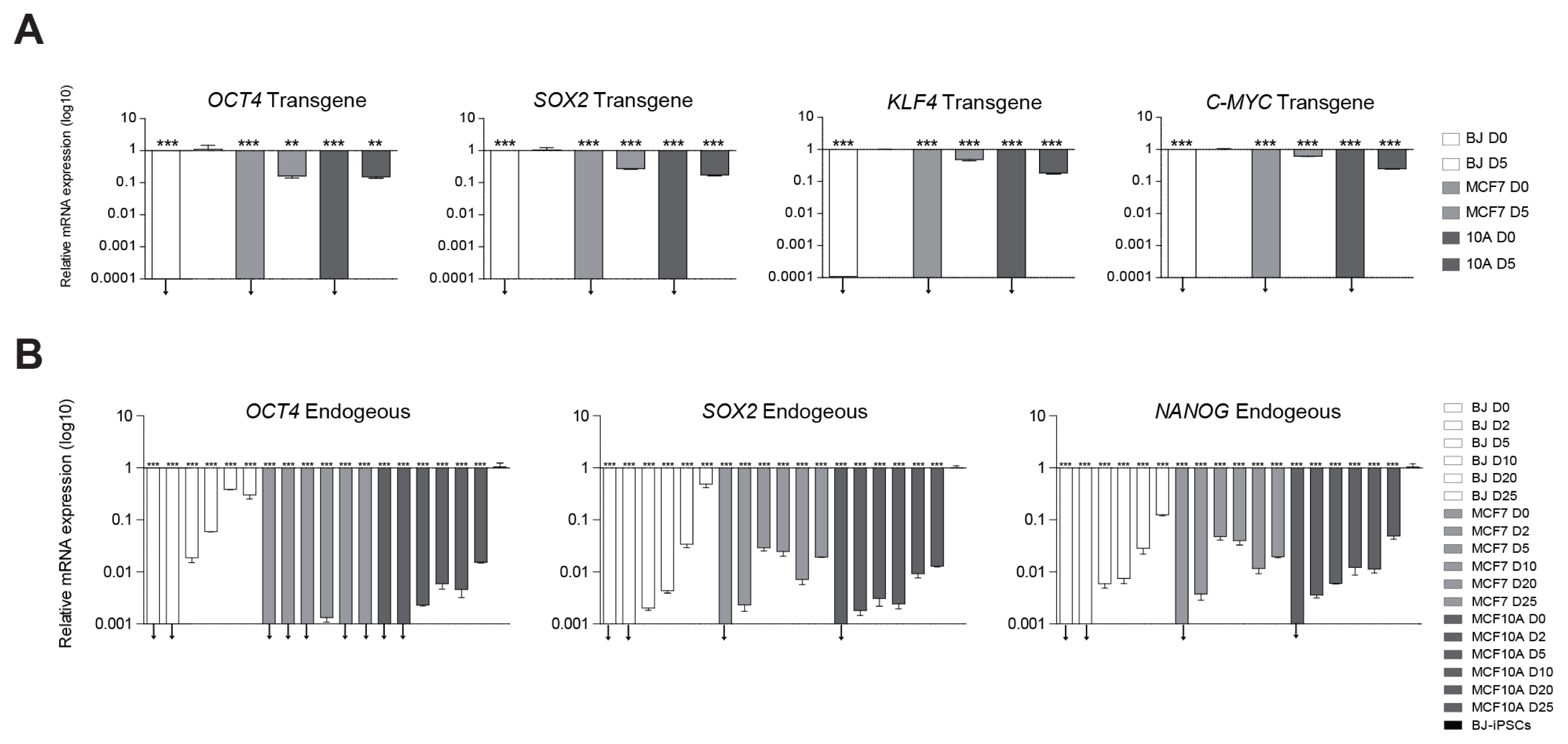
We tried induction of pluripotency in normal human fibroblasts (BJ) and both human benign (MCF10A) and malignant (MCF7) breast cancer cell lines using a classical retroviral reprogramming method. We conducted RNA-sequencing analysis to compare the transcriptome of the three cell lines at early stage of reprogramming.
RESULTS
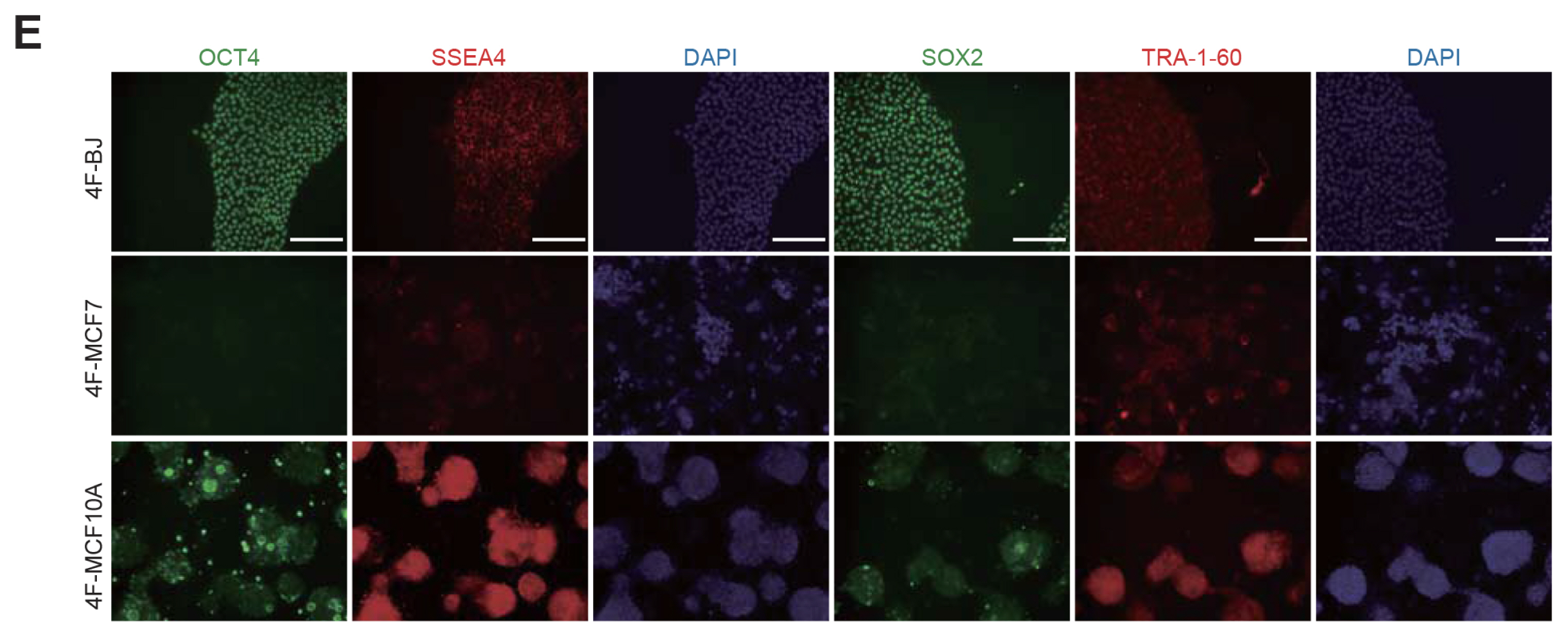
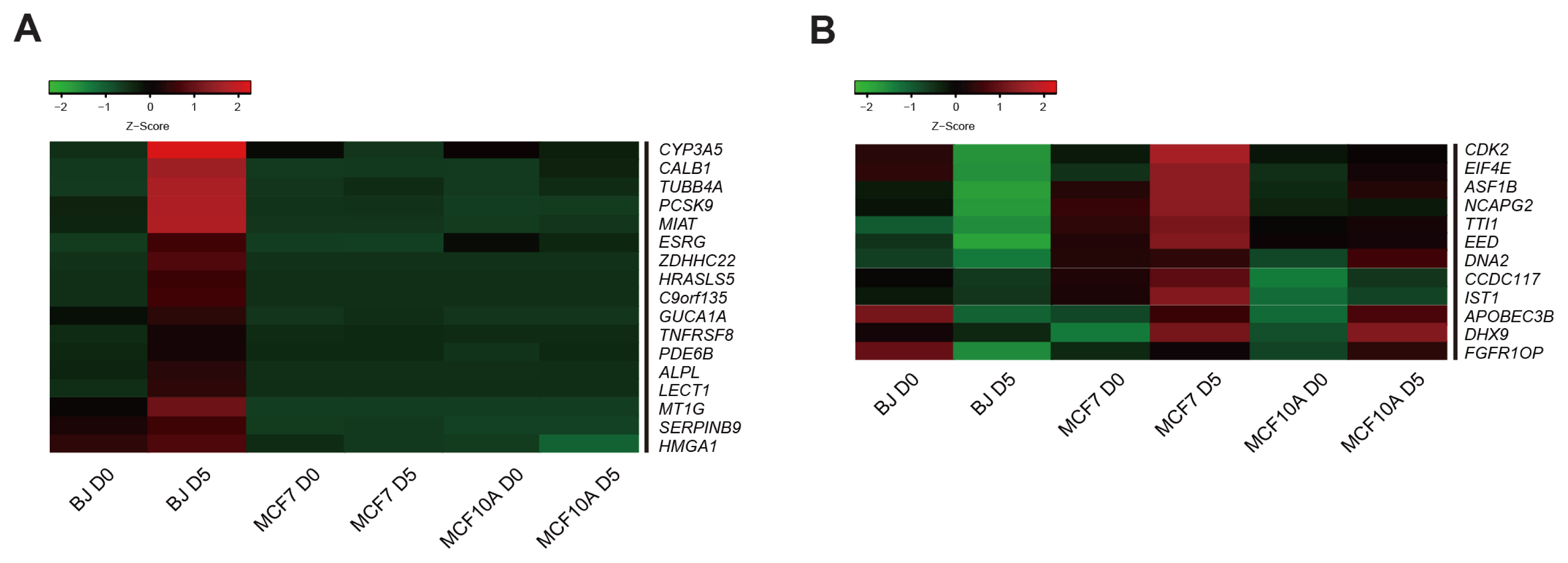
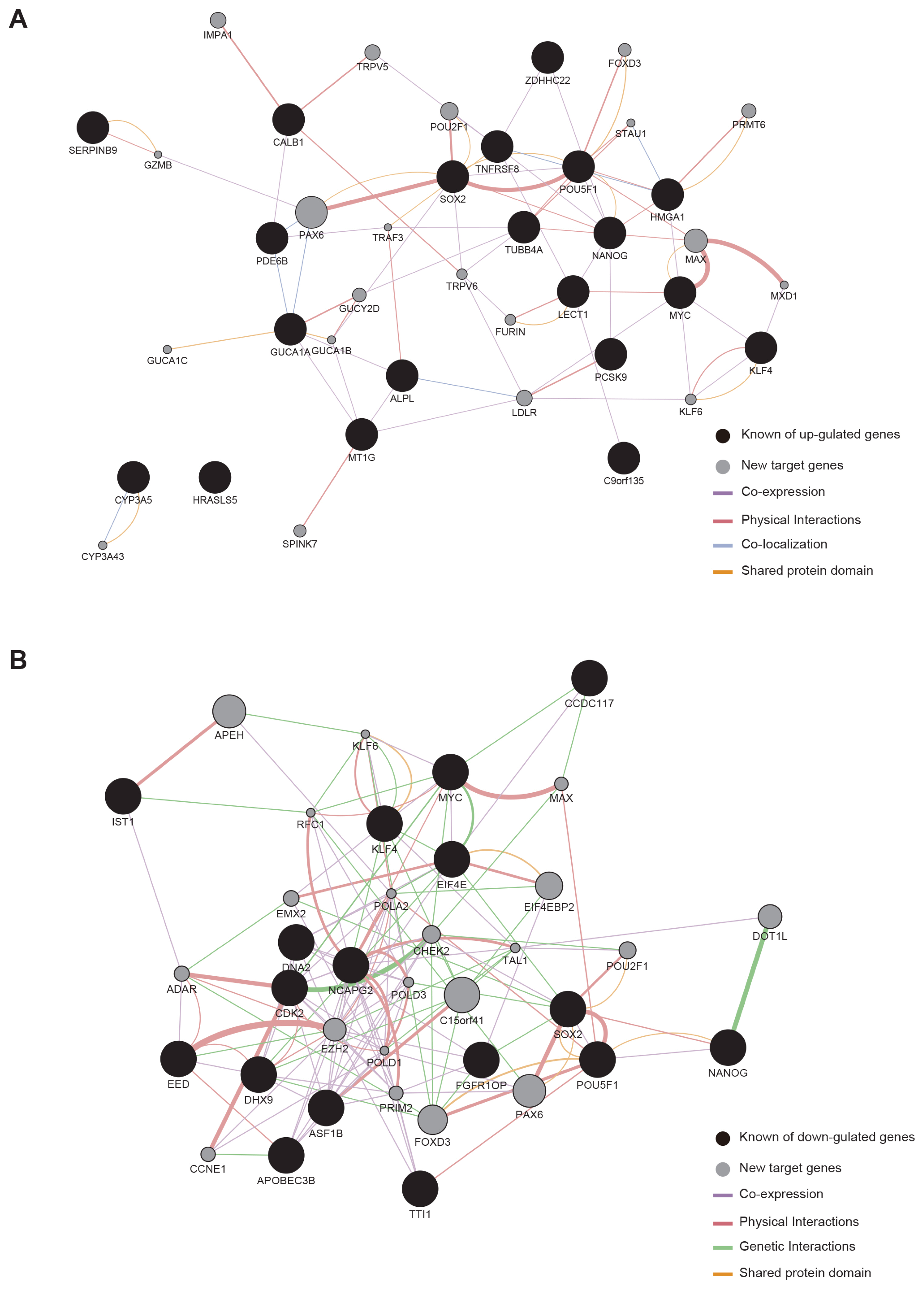
We could generate iPSCs from BJ, whereas we were unable to obtain iPSCs from cancer cell lines. To address the underlying mechanism of limited reprogramming in cancer cells, we identified 29 the candidate barrier genes based on RNA-sequencing data. In addition, we found 40 their target genes using Cytoscape software.
CONCLUSIONS
Our data suggest that these genes might one of the roadblock for cancer cell reprogramming. Furthermore, we provide new insights into application of iPSCs technology in cancer cell field for therapeutic purposes.
Keyword
MeSH Terms
Figure
Reference
-
References
1. Mair W. How normal cells can win the battle for survival against cancer cells. PLoS Biol. 2010; 8:e1000423. DOI: 10.1371/journal.pbio.1000423. PMID: 20644641. PMCID: PMC2903588.
Article2. Qu Y, Han B, Yu Y, Yao W, Bose S, Karlan BY, Giuliano AE, Cui X. Evaluation of MCF10A as a reliable model for normal human mammary epithelial cells. PLoS One. 2015; 10:e0131285. DOI: 10.1371/journal.pone.0131285. PMID: 26147507. PMCID: PMC4493126.
Article3. Osborne CK, Hobbs K, Trent JM. Biological differences among MCF-7 human breast cancer cell lines from different laboratories. Breast Cancer Res Treat. 1987; 9:111–121. DOI: 10.1007/BF01807363. PMID: 3620713.
Article4. Dai X, Cheng H, Bai Z, Li J. Breast cancer cell line classification and its relevance with breast tumor subtyping. J Cancer. 2017; 8:3131–3141. DOI: 10.7150/jca.18457. PMID: 29158785. PMCID: PMC5665029.
Article5. Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007; 318:1917–1920. DOI: 10.1126/science.1151526. PMID: 18029452.
Article6. Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007; 131:861–872. DOI: 10.1016/j.cell.2007.11.019. PMID: 18035408.
Article7. Pan XY, Tsai MH, Wuputra K, Ku CC, Lin WH, Lin YC, Kishikawa S, Noguchi M, Saito S, Lin CS, Yokoyama KK. Application of cancer cell reprogramming technology to human cancer research. Anticancer Res. 2017; 37:3367–3377. DOI: 10.21873/anticanres.11703. PMID: 28668824.
Article8. Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004; 4:143–153. DOI: 10.1038/nrc1279. PMID: 14732866.
Article9. Semi K, Matsuda Y, Ohnishi K, Yamada Y. Cellular reprogramming and cancer development. Int J Cancer. 2013; 132:1240–1248. DOI: 10.1002/ijc.27963. PMID: 23180619.
Article10. Bernhardt M, Novak D, Assenov Y, Orouji E, Knappe N, Weina K, Reith M, Larribere L, Gebhardt C, Plass C, Umansky V, Utikal J. Melanoma-derived iPCCs show differential tumorigenicity and therapy response. Stem Cell Reports. 2017; 8:1379–1391. DOI: 10.1016/j.stemcr.2017.03.007. PMID: 28392221. PMCID: PMC5425615.
Article11. Noguchi K, Eguchi H, Konno M, Kawamoto K, Nishida N, Koseki J, Wada H, Marubashi S, Nagano H, Doki Y, Mori M, Ishii H. Susceptibility of pancreatic cancer stem cells to reprogramming. Cancer Sci. 2015; 106:1182–1187. DOI: 10.1111/cas.12734. PMID: 26298849. PMCID: PMC4582987.
Article12. Miyoshi N, Ishii H, Nagai K, Hoshino H, Mimori K, Tanaka F, Nagano H, Sekimoto M, Doki Y, Mori M. Defined factors induce reprogramming of gastrointestinal cancer cells. Proc Natl Acad Sci U S A. 2010; 107:40–45. DOI: 10.1073/pnas.0912407107. PMCID: PMC2806714.
Article13. Iskender B, Izgi K, Canatan H. Reprogramming bladder cancer cells for studying cancer initiation and progression. Tumour Biol. 2016; 37:13237–13245. DOI: 10.1007/s13277-016-5226-4. PMID: 27456363.
Article14. Zhao H, Davies TJ, Ning J, Chang Y, Sachamitr P, Sattler S, Fairchild PJ, Huang FP. A highly optimized protocol for reprogramming cancer cells to pluripotency using nonviral plasmid vectors. Cell Reprogram. 2015; 17:7–18. DOI: 10.1089/cell.2014.0046. PMID: 25549177. PMCID: PMC4312798.
Article15. Corominas-Faja B, Cufí S, Oliveras-Ferraros C, Cuyàs E, López-Bonet E, Lupu R, Alarcón T, Vellon L, Iglesias JM, Leis O, Martín ÁG, Vazquez-Martin A, Menendez JA. Nuclear reprogramming of luminal-like breast cancer cells generates Sox2-overexpressing cancer stem-like cellular states harboring transcriptional activation of the mTOR pathway. Cell Cycle. 2013; 12:3109–3124. DOI: 10.4161/cc.26173. PMID: 23974095. PMCID: PMC3875684.
Article16. Mathieu J, Zhang Z, Zhou W, Wang AJ, Heddleston JM, Pinna CM, Hubaud A, Stadler B, Choi M, Bar M, Tewari M, Liu A, Vessella R, Rostomily R, Born D, Horwitz M, Ware C, Blau CA, Cleary MA, Rich JN, Ruohola-Baker H. HIF induces human embryonic stem cell markers in cancer cells. Cancer Res. 2011; 71:4640–4652. DOI: 10.1158/0008-5472.CAN-10-3320. PMID: 21712410. PMCID: PMC3129496.
Article17. Nishi M, Sakai Y, Akutsu H, Nagashima Y, Quinn G, Masui S, Kimura H, Perrem K, Umezawa A, Yamamoto N, Lee SW, Ryo A. Induction of cells with cancer stem cell properties from nontumorigenic human mammary epithelial cells by defined reprogramming factors. Oncogene. 2014; 33:643–652. DOI: 10.1038/onc.2012.614. PMID: 23318426. PMCID: PMC4697746.
Article18. Kim J, Hoffman JP, Alpaugh RK, Rhim AD, Reichert M, Stanger BZ, Furth EE, Sepulveda AR, Yuan CX, Won KJ, Donahue G, Sands J, Gumbs AA, Zaret KS. An iPSC line from human pancreatic ductal adenocarcinoma undergoes early to invasive stages of pancreatic cancer progression. Cell Rep. 2013; 3:2088–2099. DOI: 10.1016/j.celrep.2013.05.036. PMID: 23791528. PMCID: PMC3726210.
Article19. Izgi K, Canatan H, Iskender B. Current status in cancer cell reprogramming and its clinical implications. J Cancer Res Clin Oncol. 2017; 143:371–383. DOI: 10.1007/s00432-016-2258-5. PMID: 27620745.
Article20. Lin SL, Chang DC, Chang-Lin S, Lin CH, Wu DT, Chen DT, Ying SY. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA. 2008; 14:2115– 2124. DOI: 10.1261/rna.1162708. PMID: 18755840. PMCID: PMC2553732.
Article21. Zhang X, Cruz FD, Terry M, Remotti F, Matushansky I. Terminal differentiation and loss of tumorigenicity of human cancers via pluripotency-based reprogramming. Oncogene. 2013; 32:2249–2260. 2260.e1–e21. DOI: 10.1038/onc.2012.237. PMID: 22777357. PMCID: PMC3470785.
Article22. Liu Z, Che P, Mercado JJ, Hackney JR, Friedman GK, Zhang C, You Z, Zhao X, Ding Q, Kim K, Li H, Liu X, Markert JM, Nabors B, Gillespie GY, Zhao R, Han X. Characterization of iPSCs derived from low grade gliomas revealed early regional chromosomal amplifications during gliomagenesis. J Neurooncol. 2019; 141:289–301. DOI: 10.1007/s11060-018-03047-1. PMID: 30460631. PMCID: PMC6344247.
Article23. Jiang H, Lei R, Ding SW, Zhu S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing pairedend reads. BMC Bioinformatics. 2014; 15:182. DOI: 10.1186/1471-2105-15-182. PMID: 24925680. PMCID: PMC4074385.
Article24. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. DOI: 10.1093/bioinformatics/bts635. PMID: 23104886. PMCID: PMC3530905.
Article25. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010; 28:511–515. DOI: 10.1038/nbt.1621. PMID: 20436464. PMCID: PMC3146043.
Article26. Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013; 31:46–53. DOI: 10.1038/nbt.2450. PMID: 23222703. PMCID: PMC3869392.
Article27. Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S, Pico AR, Bader GD, Ideker T. A travel guide to Cytoscape plugins. Nat Methods. 2012; 9:1069–1076. DOI: 10.1038/nmeth.2212. PMID: 23132118. PMCID: PMC3649846.
Article28. Kuno A, Nishimura K, Takahashi S. Time-course transcriptome analysis of human cellular reprogramming from multiple cell types reveals the drastic change occurs between the mid phase and the late phase. BMC Genomics. 2018; 19:9. DOI: 10.1186/s12864-017-4389-8. PMID: 29298685. PMCID: PMC5753469.
Article29. Cacchiarelli D, Trapnell C, Ziller MJ, Soumillon M, Cesana M, Karnik R, Donaghey J, Smith ZD, Ratanasirintrawoot S, Zhang X, Ho Sui SJ, Wu Z, Akopian V, Gifford CA, Doench J, Rinn JL, Daley GQ, Meissner A, Lander ES, Mikkelsen TS. Integrative analyses of human reprogramming reveal dynamic nature of induced pluripotency. Cell. 2015; 162:412–424. DOI: 10.1016/j.cell.2015.06.016. PMID: 26186193. PMCID: PMC4511597.
Article30. Studer L, Vera E, Cornacchia D. Programming and reprogramming cellular age in the era of induced pluripotency. Cell Stem Cell. 2015; 16:591–600. DOI: 10.1016/j.stem.2015.05.004. PMID: 26046759. PMCID: PMC4508309.
Article31. Loh YH, Hartung O, Li H, Guo C, Sahalie JM, Manos PD, Urbach A, Heffner GC, Grskovic M, Vigneault F, Lensch MW, Park IH, Agarwal S, Church GM, Collins JJ, Irion S, Daley GQ. Reprogramming of T cells from human peripheral blood. Cell Stem Cell. 2010; 7:15–19. DOI: 10.1016/j.stem.2010.06.004. PMID: 20621044. PMCID: PMC2913590.
Article32. Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F, Vassena R, Bilić J, Pekarik V, Tiscornia G, Edel M, Boué S, Izpisúa Belmonte JC. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol. 2008; 26:1276–1284. DOI: 10.1038/nbt.1503. PMID: 18931654.
Article33. Utikal J, Maherali N, Kulalert W, Hochedlinger K. Sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells. J Cell Sci. 2009; 122(Pt 19):3502–3510. DOI: 10.1242/jcs.054783. PMID: 19723802. PMCID: PMC2746132.
Article34. Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005; 122:947–956. DOI: 10.1016/j.cell.2005.08.020. PMID: 16153702. PMCID: PMC3006442.
Article35. Veland N, Hardikar S, Zhong Y, Gayatri S, Dan J, Strahl BD, Rothbart SB, Bedford MT, Chen T. The arginine methyltransferase PRMT6 regulates DNA methylation and contributes to global DNA hypomethylation in cancer. Cell Rep. 2017; 21:3390–3397. DOI: 10.1016/j.celrep.2017.11.082. PMID: 29262320. PMCID: PMC5753604.
Article36. Lafita-Navarro MC, Blanco R, Mata-Garrido J, Liaño-Pons J, Tapia O, García-Gutiérrez L, García-Alegría E, Berciano MT, Lafarga M, León J. MXD1 localizes in the nucleolus, binds UBF and impairs rRNA synthesis. Oncotarget. 2016; 7:69536–69548. DOI: 10.18632/oncotarget.11766. PMID: 27588501. PMCID: PMC5342496.
Article37. Sun F, Chan E, Wu Z, Yang X, Marquez VE, Yu Q. Combinatorial pharmacologic approaches target EZH2-mediated gene repression in breast cancer cells. Mol Cancer Ther. 2009; 8:3191–3202. DOI: 10.1158/1535-7163.MCT-09-0479. PMID: 19934278. PMCID: PMC2794891.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Inhibition of Class I Histone Deacetylase Enhances Self-Reprogramming of Spermatogonial Stem Cells into Pluripotent Stem Cells
- Disease-specific pluripotent stem cells
- Modeling of Human Genetic Diseases Via Cellular Reprogramming
- Establishment of Hepatocellular Cancer Induced Pluripotent Stem Cells Using a Reprogramming Technique
- Induced Pluripotent Stem Cells: Next Generation Stem Cells to Clinical Applications