Healthc Inform Res.
2019 Oct;25(4):283-288. 10.4258/hir.2019.25.4.283.
Stacking Ensemble Technique for Classifying Breast Cancer
- Affiliations
-
- 1Department of IT Convergence Engineering, Gachon University, Seongnam, Korea.
- 2Department of Computer Engineering, Gachon University, Seongnam, Korea. lyh@gachon.ac.kr
- KMID: 2462226
- DOI: http://doi.org/10.4258/hir.2019.25.4.283
Abstract
OBJECTIVES
Breast cancer is the second most common cancer among Korean women. Because breast cancer is strongly associated with negative emotional and physical changes, early detection and treatment of breast cancer are very important. As a supporting tool for classifying breast cancer, we tried to identify the best meta-learner model in a stacking ensemble when the same machine learning models for the base learner and meta-learner are used.
METHODS
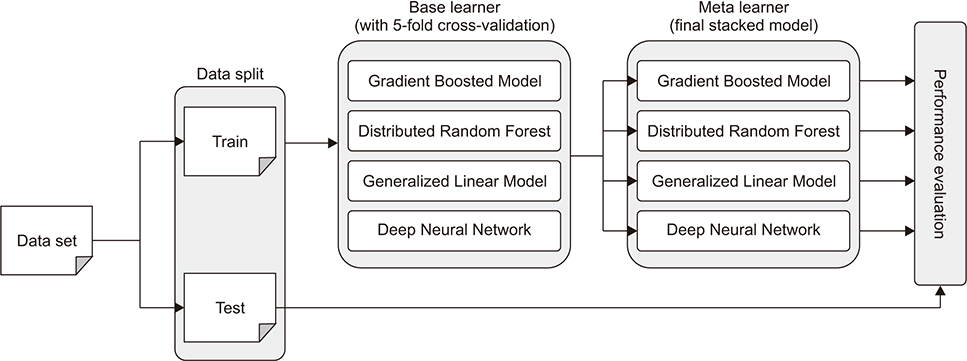
We used machine learning models, such as the gradient boosted model, distributed random forest, generalized linear model, and deep neural network in a stacking ensemble. These models were used to construct a base learner, and each of them was used as a meta-learner again. Then, we compared the performance of machine learning models in the meta-learner to determine the best meta-learner model in the stacking ensemble.
RESULTS
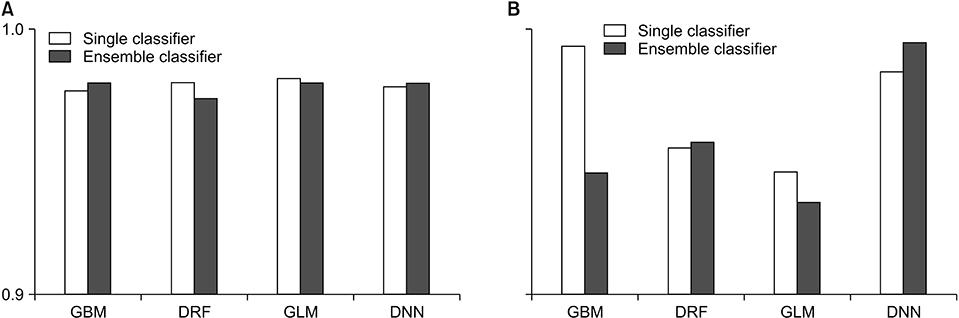
Experimental results showed that using the GBM as a meta-learner led to higher accuracy than that achieved with any other model for breast cancer data and using the GLM as a meta learner led to low root-mean-squared error for both sets of breast cancer data.
CONCLUSIONS
We compared the performance of every meta-learner model in a stacking ensemble as a supporting tool for classifying breast cancer. The study showed that using specific models as a metalearner resulted in better performance than single classifiers, and using GBM and GLM as a meta-learner is appropriate as a supporting tool for classifying breast cancer data.
MeSH Terms
Figure
Reference
-
1. Statistics Korea. Causes of Death Statistics in 2016 [Internet]. Daejeon, Korea: Statistics Korea;c2019. cited at 2018 Sep 10. Available from: http://kostat.go.kr/portal/eng/pressReleases/1/index.board?bmode=read&aSeq=363695.2. National Cancer Center. National Cancer Registration and Statistics in Korea 2015 [Internet]. Goyang, Korea: National Cancer Center;c2019. cited at 2018 Sep 10. Available from: http://ncc.re.kr/main.ncc?uri=english/sub04_Statistics.3. Choi BJ, Park JH, Choe BM, Han SH, Kim SH. Factors influencing anxiety and depression in breast cancer patients treated with surgery. J Korean Soc Biol Ther Psychiatry. 2011; 17(1):87–95.4. Nam SJ. Screening and diagnosis for breast cancers. J Korean Med Assoc. 2009; 52(10):946–951.
Article5. Jung SS, You YK, Park CH, Kim IC. Recent trends of breast cancer treatment in Korea. J Korean Surg Soc. 1991; 41(6):717–726.6. Kim DD, Kim JH, Choi KW. The clinicopathologic factors affecting the false negativity of fine needle aspiration cytology (FNAC) in breast Cancer. J Korean Surg Soc. 2002; 62(5):403–407.7. Layfield LJ, Glasgow BJ, Cramer H. Fine-needle aspiration in the management of breast masses. Pathol Annu. 1989; 24 Pt 2:23–62.8. Oh BH, Park YS, Sung CW, Kim CS. Diagnostic value of ultrasound-guided fine needle aspiration cytology by a endocrine surgeon. Korean J Endocr Surg. 2008; 8(3):189–193.
Article9. Hong SW. Fine needle aspiration cytology of thyroid follicular proliferative lesions. Korean J Endocr Surg. 2008; 8(3):159–166.
Article10. Cho SJ, Kang SH. Industrial applications of machine learning (artificial intelligence). IE Mag. 2016; 23(2):34–38.11. van der Laan MJ, Polley EC, Hubbard AE. Super learner. Stat Appl Genet Mol Biol. 2007; 6:Article25.
Article12. Lim JS, Oh YS, Lim DH. Bagging support vector machine for improving breast cancer classification. J Health Info Stat. 2014; 39(1):15–24.13. Krawczyk B, Galar M, Jelen L, Herrera F. Evolutionary undersampling boosting for imbalanced classification of breast cancer malignancy. Appl Soft Comput. 2016; 38:714–726.
Article14. Huang MW, Chen CW, Lin WC, Ke SW, Tsai CF. SVM and SVM Ensembles in Breast Cancer Prediction. PLoS One. 2017; 12(1):e0161501.
Article15. Nagi S, Bhattacharyya DK. Classification of microarray cancer data using ensemble approach. Netw Model Anal Health Inform Bioinform. 2013; 2(3):159–173.
Article16. UCI Machine Learning Repository [Internet]. Oakland (CA): University of California, Center for Machine Learning and Intelligent Systems;c2018. cited at 2018 Aug 10. Available from: http://archive.ics.uci.edu/ml.17. H2O documentation overview [Internet]. [place unknown]: H2O.AI;2019. cited 2018 Aug 10. Available from: http://docs.h2o.ai/h2o/latest-stable/h2o-docs/index.html.18. Park JH, Jung YS, Jung Y. Factors influencing posttraumatic growth in survivors of breast cancer. J Korean Acad Nurs. 2016; 46(3):454–462.
Article19. Montazeri M, Montazeri M, Montazeri M, Beigzadeh A. Machine learning models in breast cancer survival prediction. Technol Health Care. 2016; 24(1):31–42.
Article20. Hsieh SL, Hsieh SH, Cheng PH, Chen CH, Hsu KP, Lee IS, et al. Design ensemble machine learning model for breast cancer diagnosis. J Med Syst. 2012; 36(5):2841–2847.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Highligts of 28th Annual Meeting of San Antonio Breast Cancer Symposium
- Surgery of Breast Cancer during the Last 5 Years: More Sophisticated and Specialized?
- Modified Five Field Technique for Primary and Postop Breast Cancer Irradiation
- Mono- and Combination Chemotherapy for Metastatic Breast Cancer: An Increamental Step Forward
- Aromtase Inhibitors in Breast Cancer