Korean J Physiol Pharmacol.
2019 Nov;23(6):539-547. 10.4196/kjpp.2019.23.6.539.
Deficiency of Anoctamin 5/TMEM16E causes nuclear positioning defect and impairs Ca²⺠signaling of differentiated C2C12 myotubes
- Affiliations
-
- 1Department of Physiology, Sungkyunkwan University School of Medicine, Suwon 16419, Korea. tongmkang@skku.edu
- KMID: 2461047
- DOI: http://doi.org/10.4196/kjpp.2019.23.6.539
Abstract
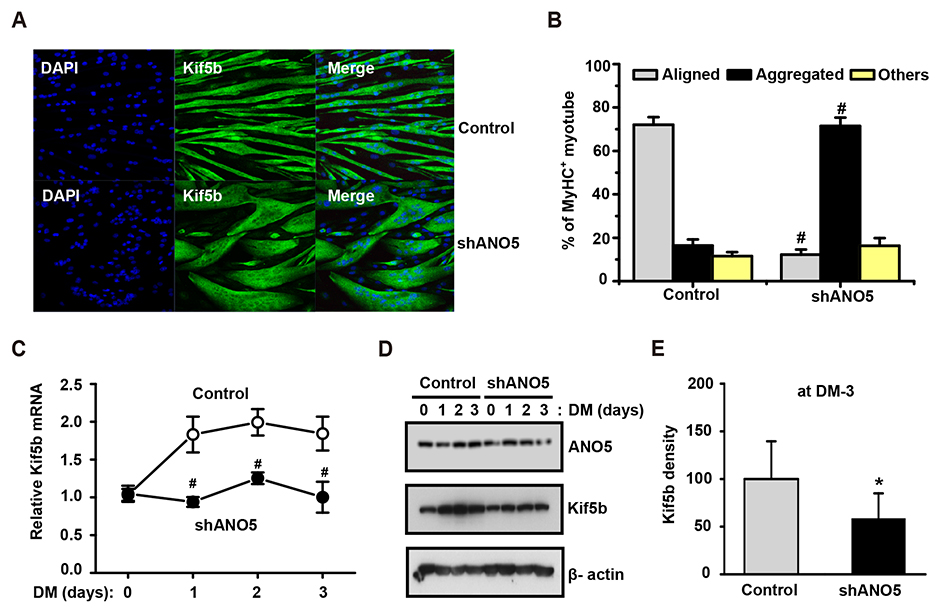
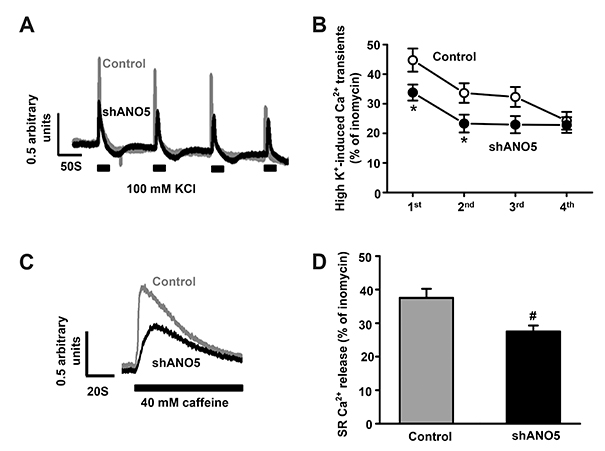
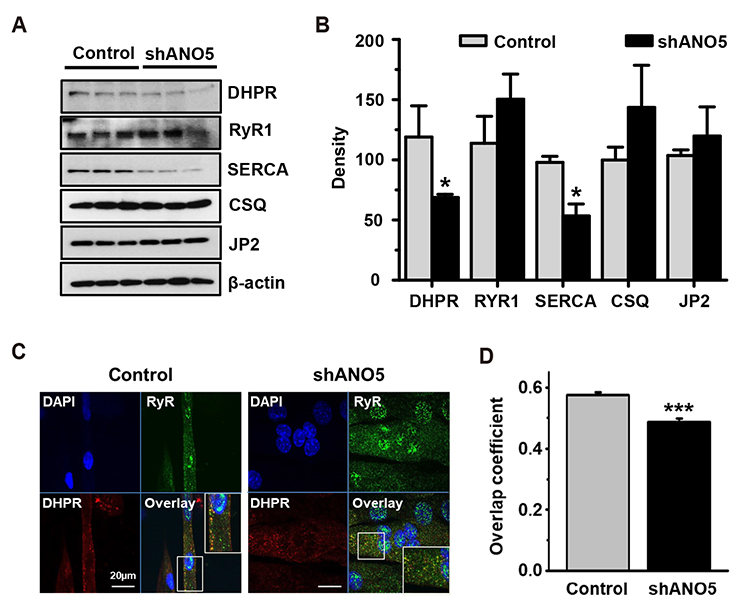
- Anoctamin 5 (ANO5)/TMEM16E belongs to a member of the ANO/TMEM16 family member of anion channels. However, it is a matter of debate whether ANO5 functions as a genuine plasma membrane chloride channel. It has been recognized that mutations in the ANO5 gene cause many skeletal muscle diseases such as limb girdle muscular dystrophy type 2L (LGMD2L) and Miyoshi muscular dystrophy type 3 (MMD3) in human. However, the molecular mechanisms of the skeletal myopathies caused by ANO5 defects are poorly understood. To understand the role of ANO5 in skeletal muscle development and function, we silenced the ANO5 gene in C2C12 myoblasts and evaluated whether it impairs myogenesis and myotube function. ANO5 knockdown (ANO5-KD) by shRNA resulted in clustered or aggregated nuclei at the body of myotubes without affecting differentiation or myotube formation. Nuclear positioning defect of ANO5-KD myotubes was accompanied with reduced expression of Kif5b protein, a kinesin-related motor protein that controls nuclear transport during myogenesis. ANO5-KD impaired depolarization-induced [Ca²âº]i transient and reduced sarcoplasmic reticulum (SR) Ca²âº storage. ANO5-KD resulted in reduced protein expression of the dihydropyridine receptor (DHPR) and SR Ca²âº-ATPase subtype 1. In addition, ANO5-KD compromised co-localization between DHPR and ryanodine receptor subtype 1. It is concluded that ANO5-KD causes nuclear positioning defect by reduction of Kif5b expression, and compromises Ca²âº signaling by downregulating the expression of DHPR and SERCA proteins.
Keyword
MeSH Terms
-
Active Transport, Cell Nucleus
Calcium Channels, L-Type
Cell Membrane
Chloride Channels
Humans
Muscle Development
Muscle Fibers, Skeletal*
Muscle, Skeletal
Muscular Diseases
Muscular Dystrophies
Muscular Dystrophies, Limb-Girdle
Myoblasts
RNA, Small Interfering
Ryanodine Receptor Calcium Release Channel
Sarcoplasmic Reticulum
Calcium Channels, L-Type
Chloride Channels
RNA, Small Interfering
Ryanodine Receptor Calcium Release Channel
Figure
Reference
-
1. Pedemonte N, Galietta LJ. Structure and function of TMEM16 proteins (anoctamins). Physiol Rev. 2014; 94:419–459.
Article2. Yang YD, Cho H, Koo JY, Tak MH, Cho Y, Shim WS, Park SP, Lee J, Lee B, Kim BM, Raouf R, Shin YK, Oh U. TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature. 2008; 455:1210–1215.
Article3. Stephan AB, Shum EY, Hirsh S, Cygnar KD, Reisert J, Zhao H. ANO2 is the cilial calcium-activated chloride channel that may mediate olfactory amplification. Proc Natl Acad Sci U S A. 2009; 106:11776–11781.
Article4. Tian Y, Schreiber R, Kunzelmann K. Anoctamins are a family of Ca2+-activated Cl- channels. J Cell Sci. 2012; 125(Pt 21):4991–4998.5. Duran C, Qu Z, Osunkoya AO, Cui Y, Hartzell HC. ANOs 3-7 in the anoctamin/Tmem16 Cl- channel family are intracellular proteins. Am J Physiol Cell Physiol. 2012; 302:C482–C493.
Article6. Xu J, El Refaey M, Xu L, Zhao L, Gao Y, Floyd K, Karaze T, Janssen PM, Han R. Genetic disruption of Ano5 in mice does not recapitulate human ANO5-deficient muscular dystrophy. Skelet Muscle. 2015; 5:43.
Article7. Bouquet F, Cossée M, Béhin A, Deburgrave N, Romero N, Leturcq F, Eymard B. Miyoshi-like distal myopathy with mutations in anoctamin 5 gene. Rev Neurol (Paris). 2012; 168:135–141.
Article8. Penttilä S, Palmio J, Suominen T, Raheem O, Evilä A, Muelas Gomez N, Tasca G, Waddell LB, Clarke NF, Barboi A, Hackman P, Udd B. Eight new mutations and the expanding phenotype variability in muscular dystrophy caused by ANO5. Neurology. 2012; 78:897–903.
Article9. Tsutsumi S, Kamata N, Vokes TJ, Maruoka Y, Nakakuki K, Enomoto S, Omura K, Amagasa T, Nagayama M, Saito-Ohara F, Inazawa J, Moritani M, Yamaoka T, Inoue H, Itakura M. The novel gene encoding a putative transmembrane protein is mutated in gnathodiaphyseal dysplasia (GDD). Am J Hum Genet. 2004; 74:1255–1261.
Article10. Hicks D, Sarkozy A, Muelas N, Köehler K, Huebner A, Hudson G, Chinnery PF, Barresi R, Eagle M, Polvikoski T, Bailey G, Miller J, Radunovic A, Hughes PJ, Roberts R, Krause S, Walter MC, Laval SH, Straub V, Lochmüller H, Bushby K. A founder mutation in Anoctamin 5 is a major cause of limb-girdle muscular dystrophy. Brain. 2011; 134(Pt 1):171–182.11. Little AA, McKeever PE, Gruis KL. Novel mutations in the Anoctamin 5 gene (ANO5) associated with limb-girdle muscular dystrophy 2L. Muscle Nerve. 2013; 47:287–291.
Article12. Bolduc V, Marlow G, Boycott KM, Saleki K, Inoue H, Kroon J, Itakura M, Robitaille Y, Parent L, Baas F, Mizuta K, Kamata N, Richard I, Linssen WH, Mahjneh I, de Visser M, Bashir R, Brais B. Recessive mutations in the putative calcium-activated chloride channel Anoctamin 5 cause proximal LGMD2L and distal MMD3 muscular dystrophies. Am J Hum Genet. 2010; 86:213–221.
Article13. Griffin DA, Johnson RW, Whitlock JM, Pozsgai ER, Heller KN, Grose WE, Arnold WD, Sahenk Z, Hartzell HC, Rodino-Klapac LR. Defective membrane fusion and repair in Anoctamin5-deficient muscular dystrophy. Hum Mol Genet. 2016; 25:1900–1911.14. Whitlock JM, Yu K, Cui YY, Hartzell HC. Anoctamin 5/TMEM16E facilitates muscle precursor cell fusion. J Gen Physiol. 2018; 150:1498–1509.
Article15. Gache V, Gomes ER, Cadot B. Microtubule motors involved in nuclear movement during skeletal muscle differentiation. Mol Biol Cell. 2017; 28:865–874.
Article16. Metzger T, Gache V, Xu M, Cadot B, Folker ES, Richardson BE, Gomes ER, Baylies MK. MAP and kinesin-dependent nuclear positioning is required for skeletal muscle function. Nature. 2012; 484:120–124.
Article17. Wilson MH, Holzbaur EL. Opposing microtubule motors drive robust nuclear dynamics in developing muscle cells. J Cell Sci. 2012; 125(Pt 17):4158–4169.
Article18. Wang Z, Cui J, Wong WM, Li X, Xue W, Lin R, Wang J, Wang P, Tanner JA, Cheah KS, Wu W, Huang JD. Kif5b controls the localization of myofibril components for their assembly and linkage to the myotendinous junctions. Development. 2013; 140:617–626.
Article19. Phuong TT, Yun YH, Kim SJ, Kang TM. Positive feedback control between STIM1 and NFATc3 is required for C2C12 myoblast differentiation. Biochem Biophys Res Commun. 2013; 430:722–728.
Article20. Sunadome K, Yamamoto T, Ebisuya M, Kondoh K, Sehara-Fujisawa A, Nishida E. ERK5 regulates muscle cell fusion through Klf transcription factors. Dev Cell. 2011; 20:192–205.
Article21. Chandra G, Defour A, Mamchoui K, Pandey K, Mishra S, Mouly V, Sreetama S, Mahad Ahmad M, Mahjneh I, Morizono H, Pattabiraman N, Menon AK, Jaiswal JK. Dysregulated calcium homeostasis prevents plasma membrane repair in Anoctamin 5/TMEM16Edeficient patient muscle cells. Cell Death Discov. 2019; 5:118.
Article22. Magri F, Del Bo R, D'Angelo MG, Sciacco M, Gandossini S, Govoni A, Napoli L, Ciscato P, Fortunato F, Brighina E, Bonato S, Bordoni A, Lucchini V, Corti S, Moggio M, Bresolin N, Comi GP. Frequency and characterisation of anoctamin 5 mutations in a cohort of Italian limb-girdle muscular dystrophy patients. Neuromuscul Disord. 2012; 22:934–943.
Article23. Di Zanni E, Gradogna A, Scholz-Starke J, Boccaccio A. Gain of function of TMEM16E/ANO5 scrambling activity caused by a mutation associated with gnathodiaphyseal dysplasia. Cell Mol Life Sci. 2018; 75:1657–1670.
Article24. Liu JH, König S, Michel M, Arnaudeau S, Fischer-Lougheed J, Bader CR, Bernheim L. Acceleration of human myoblast fusion by depolarization: graded Ca2+ signals involved. Development. 2003; 130:3437–3446.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Secretion of adenylate kinase 1 is required for extracellular ATP synthesis in C2C12 myotubes
- The effects of Allomyrina dichotoma larval extract on palmitate-induced insulin resistance in skeletal muscle cells
- 17Beta-estradiol Stimulates Glucose Uptake Through Estrogen Receptor and AMP-activated Protein Kinase Activation in C2C12 Myotubes
- Glycolytic and oxidative muscles under acute glucose supplementation differ in their metabolic responses to fatty acyl-CoA synthetase gene suppression
- Rg3 Improves Mitochondrial Function and the Expression of Key Genes Involved in Mitochondrial Biogenesis in C2C12 Myotubes