Ann Lab Med.
2020 Jan;40(1):27-32. 10.3343/alm.2020.40.1.27.
A Novel Mismatched PCR-Restriction Fragment Length Polymorphism Assay for Rapid Detection of gyrA and parC Mutations Associated With Fluoroquinolone Resistance in Acinetobacter baumannii
- Affiliations
-
- 1Department of Microbiology and Infectious Diseases, Nara Medical University, Nara, Japan. rnakano@naramed-u.ac.jp
- 2Department of Otolaryngology-Head and Neck Surgery, Nara Medical University, Nara, Japan.
- 3International University of Health and Welfare, Shioya Hospital, Tochigi, Japan.
- 4Department of Otolaryngology-Head and Neck Surgery, Tohoku University Graduate School of Medicine, Miyagi, Japan.
- 5Department of Microbiology and Immunology, Teikyo University School of Medicine, Tokyo, Japan.
- KMID: 2457490
- DOI: http://doi.org/10.3343/alm.2020.40.1.27
Abstract
- BACKGROUND
Mutations in the quinolone resistance-determining regions (QRDRs) of Acinetobacter baumannii DNA gyrase (gyrA) and topoisomerase IV (parC) are linked to fluoroquinolone (FQ) resistance. We developed a mismatched PCR-restriction fragment length polymorphism (RFLP) assay to detect mutations in the gyrA and parC QRDRs associated with FQ resistance in A. baumannii.
METHODS
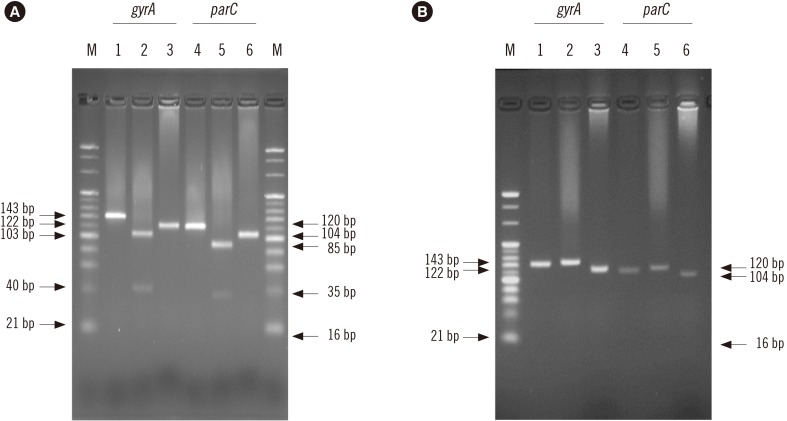
Based on the conserved sequences of A. baumannii gyrA and parC, two primer sets were designed for mismatched PCR-RFLP to detect mutations in gyrA (codons 83 and 87) and parC (codons 80 and 84) by introducing an artificial restriction enzyme cleavage site into the PCR products. This assay was evaluated using 58 A. baumannii strains and 37 other Acinetobacter strains that have been identified by RNA polymerase β-subunit gene sequence analysis.
RESULTS
PCR amplification of gyrA and parC was successful for all A. baumannii strains. In 11 FQ -susceptible strains, the gyrA and parC PCR products were digested by the selected restriction enzymes at the site containing gyrA (codons 83 and 87) and parC (codons 80 and 84). PCR products from 47 FQ-resistant strains containing mutations in gyrA and parC were not digested by the restriction enzymes at the site containing the mutation. As for the non-baumannii Acinetobacter strains, although amplification products for gyrA were obtained for 28 strains, no parC amplification product was obtained for any strain.
CONCLUSIONS
This assay specifically amplified gyrA and parC from A. baumannii and detected A. baumannii gyrA and parC mutations with FQ resistance.
Keyword
MeSH Terms
Figure
Reference
-
1. Hooper DC. Clinical applications of quinolones. Biochim Biophys Acta. 1998; 1400:45–61. PMID: 9748496.2. Kim ES, Hooper DC. Clinical importance and epidemiology of quinolone resistance. Infect Chemother. 2014; 46:226–238. PMID: 25566402.3. Dalhoff A. Resistance surveillance studies: a multifaceted problem--the fluoroquinolone example. Infection. 2012; 40:239–262. PMID: 22460782.4. Gaynes R, Edwards JR. National Nosocomial Infections Surveillance System. Overview of nosocomial infections caused by gram-negative bacilli. Clin Infect Dis. 2005; 41:848–854. PMID: 16107985.5. Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004; 39:309–317. PMID: 15306996.6. Vila J, Ruiz J, Goñi P, Marcos A, Jimenez de Anta T. Mutation in the gyrA gene of quinolone-resistant clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother. 1995; 39:1201–1203. PMID: 7625818.7. Vila J, Ruiz J, Goñi P, Jimenez de Anta T. Quinolone-resistance mutations in the topoisomerase IV parC gene of Acinetobacter baumannii. J Antimicrob Chemother. 1997; 39:757–762. PMID: 9222045.8. Wisplinghoff H, Decker M, Haefs C, Krut O, Plum G, Seifert H. Mutations in gyrA and parC associated with resistance to fluoroquinolones in epidemiologically defined clinical strains of Acinetobacter baumannii. J Antimicrob Chemother. 2003; 51:177–180. PMID: 12493807.9. Coyne S, Courvalin P, Périchon B. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob Agents Chemother. 2011; 55:947–953. PMID: 21173183.10. Correia S, Poeta P, Hébraud M, Capelo JL, Igrejas G. Mechanisms of quinolone action and resistance: where do we stand? J Med Microbiol. 2017; 66:551–559. PMID: 28504927.11. Valentine SC, Contreras D, Tan S, Real LJ, Chu S, Xu HH. Phenotypic and molecular characterization of Acinetobacter baumannii clinical isolates from nosocomial outbreaks in Los Angeles County, California. J Clin Microbiol. 2008; 46:2499–2507. PMID: 18524965.12. Ruiz J. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J Antimicrob Chemother. 2003; 51:1109–1117. PMID: 12697644.13. Hamouda A, Amyes SG. Novel gyrA and parC point mutations in two strains of Acinetobacter baumannii resistant to ciprofloxacin. J Antimicrob Chemother. 2004; 54:695–696. PMID: 15282231.14. Nakano R, Okamoto R, Nakano A, Nagano N, Abe M, Tansho-Nagakawa S, et al. Rapid assay for detecting gyrA and parC mutations associated with fluoroquinolone resistance in Enterobacteriaceae. J Microbiol Methods. 2013; 94:213–216. PMID: 23816531.15. Endo S, Yano H, Hirakata Y, Arai K, Kanamori H, Ogawa M, et al. Molecular epidemiology of carbapenem-non-susceptible Acinetobacter baumannii in Japan. J Antimicrob Chemother. 2012; 67:1623–1626. PMID: 22447879.16. Mu X, Nakano R, Nakano A, Ubagai T, Kikuchi-Ueda T, Tansho-Nagakawa S, et al. Loop-mediated isothermal amplification: Rapid and sensitive detection of the antibiotic resistance gene ISAba1-blaOXA-51-like in Acinetobacter baumannii. J Microbiol Methods. 2016; 121:36–40. PMID: 26707336.17. La Scola B, Gundi VA, Khamis A, Raoult D. Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J Clin Microbiol. 2006; 44:827–832. PMID: 16517861.18. Ministry of Health, Labour and Welfare. Ethical Guidelines for Medical and Health Research Involving Human Subjects. https://www.lifescience.mext.go.jp/bioethics/ekigaku.html.19. CLSI. Performance standards for antimicrobial susceptibility testing. CLSI supplement M100-S22. 22nd ed. Wayne, PA: Clinical and Laboratory Standards Institute;2012.20. Pournaras S, Markogiannakis A, Ikonomidis A, Kondyli L, Bethimouti K, Maniatis AN, et al. Outbreak of multiple clones of imipenem-resistant Acinetobacter baumannii isolates expressing OXA-58 carbapenemase in an intensive care unit. J Antimicrob Chemother. 2006; 57:557–561. PMID: 16431857.21. Hujer KM, Hujer AM, Endimiani A, Thomson JM, Adams MD, Goglin K, et al. Rapid determination of quinolone resistance in Acinetobacter spp. J Clin Microbiol. 2009; 47:1436–1442. PMID: 19297590.22. Haliassos A, Chomel JC, Tesson L, Baudis M, Kruh J, Kaplan JC, et al. Modification of enzymatically amplified DNA for the detection of point mutations. Nucleic Acids Res. 1989; 17:3606. PMID: 2726503.23. Deccache Y, Irenge LM, Savov E, Ariciuc M, Macovei A, Trifonova A, et al. Development of a pyrosequencing assay for rapid assessment of quinolone resistance in Acinetobacter baumannii isolates. J Microbiol Methods. 2011; 86:115–118. PMID: 21514328.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Alterations of
gyrA, gyrB , andparC and Activity of Efflux Pump in Fluoroquinolone-resistantAcinetobacter baumannii - Molecular Investigation of Quinolone Resistance of Quinolone Resistance-Determining Region in
Streptococcus pneumoniae Strains Isolated from Iran Using Polymerase Chain Reaction–Restriction Fragment Length Polymorphism Method - Genospecies Classification of Acinetobacter calcoaceticus-Acinetobacter baumannii Complex by Restriction Fragment Length Polymorphism
- Genetic Basis of Multidrug-resistant Acinetobacter baumannii Clinical Isolates from Three University Hospitals in Chungcheong Province, Korea
- Antimicrobial Resistance Determinants in Imipenem-nonsusceptible Acinetobacter calcoaceticus-baumannii Complex Isolated in Daejeon, Korea
- Alterations of