J Bacteriol Virol.
2019 Jun;49(2):53-58. 10.4167/jbv.2019.49.2.53.
Association Between Toxin-antitoxin Systems on Plasmids and Persister Formation in CTX-15-producing Klebsiella pneumoniae ST11 Isolates
- Affiliations
-
- 1Department of Molecular Cell Biology, Sungkyunkwan University School of Medicine, Suwon 16419, South Korea. ksko@skku.edu
- KMID: 2452046
- DOI: http://doi.org/10.4167/jbv.2019.49.2.53
Abstract
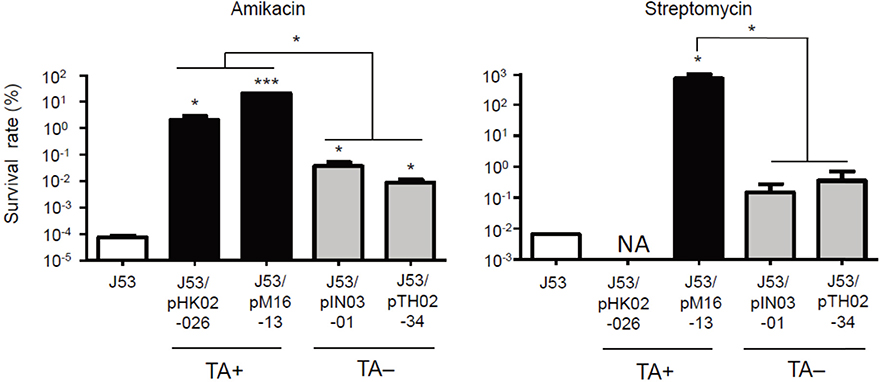
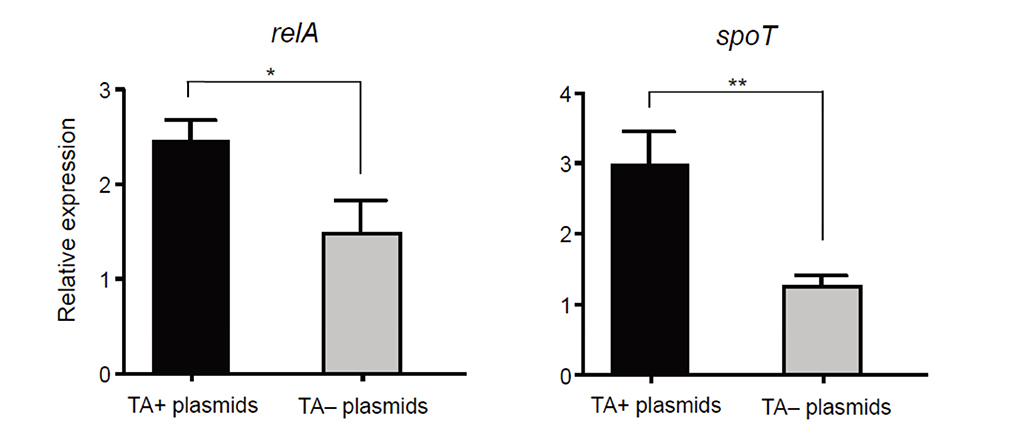
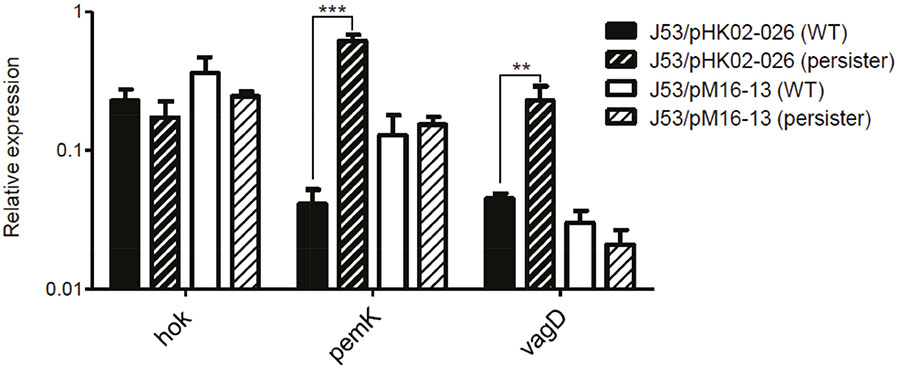
- We investigated the effect of toxin-antitoxin (TA) systems in bla(CTX-M-15)-bearing plasmids of Klebsiella pneumoniae on persister formation. The persister formation rate was notably high in transconjugants in plasmids bearing TA system than the transconjugants in plasmids bearing no TA systems. Activation of relA and spoT expression was higher in transconjugants with plasmids bearing TA systems. Thus, TA systems in plasmids may contribute to the maintenance of bla(CTX-M-15)-bearing plasmids and host survival via persister formation.
MeSH Terms
Figure
Reference
-
1. Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017; 41:252–275.
Article2. Mathers AJ, Peirano G, Pitout JD. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev. 2015; 28:565–591.
Article3. Ko KS, Lee JY, Baek JY, Suh JY, Lee MY, Choi JY, et al. Predominance of an ST11 extended-spectrum beta-lactamase-producing Klebsiella pneumoniae clone causing bacteraemia and urinary tract infections in Korea. J Med Microbiol. 2010; 59:822–828.
Article4. Zhao WH, Hu ZQ. Epidemiology and genetics of CTX-M extended-spectrum β-lactamases in Gram-negative bacteria. Crit Rev Microbiol. 2013; 39:79–101.
Article5. Shin J, Ko KS. Comparative study of genotype and virulence in CTX-M-producing and non-extended-spectrum-β-lactamase-producing Klebsiella pneumoniae isolates. Antimicrob Agents Chemother. 2014; 58:2463–2467.
Article6. Shin J, Ko KS. Effect of plasmids harbouring blaCTX-M on the virulence and fitness of Escherichia coli ST131 isolates. Int J Antimicrob Agents. 2015; 46:214–218.
Article7. Woodford N, Carattoli A, Karisik E, Underwood A, Ellington MJ, Livermore DM. Complete nucleotide sequences of plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrob Agents Chemother. 2009; 53:4472–4482.
Article8. Doumith M, Findlay J, Hirani H, Hopkins KL, Livermore DM, Dodgson A, et al. Major role of pKpQIL-like plasmids in the early dissemination of KPC-type carbapenemases in the UK. J Antimicrob Chemother. 2017; 72:2241–2248.
Article9. Page R, Peti W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat Chem Biol. 2016; 12:208–214.
Article10. Lewis K. Persister cells. Annu Rev Microbiol. 2010; 64:357–372.
Article11. Michiels JE, Van den Bergh B, Verstraeten N, Michiels J. Molecular mechanisms and clinical implications of bacterial persistence. Drug Resist Updat. 2016; 29:76–89.
Article12. Chung ES, Wi YM, Ko KS. Variation in formation of persister cells against colistin in Acinetobacter baumannii isolates and its relationship with treatment failure. J Antimicrob Chemother. 2017; 72:2133–2135.
Article13. Barrett TC, MokWWK , Murawski AM, Brynildsen MP. Enhanced antibiotic resistance development from fluoroquinolone persisters after a single exposure to antibiotic. Nat Commun. 2019; 10:1177.
Article14. Kim SY, Ko KS. Diverse plasmids harboring blaCTX-M-15 in Klebsiella pneumoniae ST11 isolates from several Asian countries. Microb Drug Resist. 2019; 25:227–232.
Article15. Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing: 27th informational supplement, M100-S27. Wayne, PA: CLSI;2017.16. Bhargava N, Sharma P, Capalash N. Pyocyanin stimulates quorum sensing-mediated tolerance to oxidative stress and increases persister cell populations in Acinetobacter baumannii. Infect Immun. 2014; 82:3417–3425.
Article17. Lee JY, Chung ES, Na IY, Kim H, Shin D, Ko KS. Development of colistin resistance in pmrA-, phoP-, parR-, and cprR-inactivated mutants of Pseudomonas aeruginosa. J Antimicrob Chemother. 2014; 69:2966–2971.18. Fernández-García L, Blasco L, Lopez M, Bou G, García-Contreras R, Wood T, et al. Toxin-antitoxin systems in clinical pathogens. Toxins (Basel). 2016; 8:E227.
Article19. Kamarthapu V, Epshtein V, Benjamin B, Proshkin S, Mironov A, Cashel M, et al. ppGpp couples transcription to DNA repair in E. coli. Science. 2016; 352:993–996.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- In Vitro Susceptibility of piperacillin/tazobactam Against extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae
- Prevalence and antimicrobial resistance of Klebsiella species isolated from clinically ill companion animals
- Prevalence of CTX-M-type Extended-Spectrum beta-Lactamase-Producing Esherichia coli and Klebsiella pneumoniae Isolates in Korea
- Prevalence of CTX-M Extended-spectrum Beta-lactamases in Clinical Isolates of Enterobacteriaceae in Korea
- Comparisons of CTX-M-Producing Escherichia coli Isolates from Humans and Animals in South Korea