J Clin Neurol.
2019 Jan;15(1):102-107. 10.3988/jcn.2019.15.1.102.
Effect of Single-Nucleotide Polymorphisms on Decline of Dopamine Transporter Availability in Parkinson's Disease
- Affiliations
-
- 1Department of Nuclear Medicine, Biomedical Research Institute, Pusan National University Hospital, Busan, Korea. injkim@pusan.ac.kr, ilikechopin@me.com
- 2Department of Neurosurgery, Biomedical Research Institute, Pusan National University Hospital, Busan, Korea.
- 3Department of Neurology, Biomedical Research Institute, Pusan National University Hospital, Busan, Korea. mslayer9@gmail.com
- 4Department of Nuclear Medicine and Research Institute for Convergence of Biomedical Science and Technology, Pusan National University Yangsan Hospital, Yangsan, Korea.
- KMID: 2451152
- DOI: http://doi.org/10.3988/jcn.2019.15.1.102
Abstract
- BACKGROUND AND PURPOSE
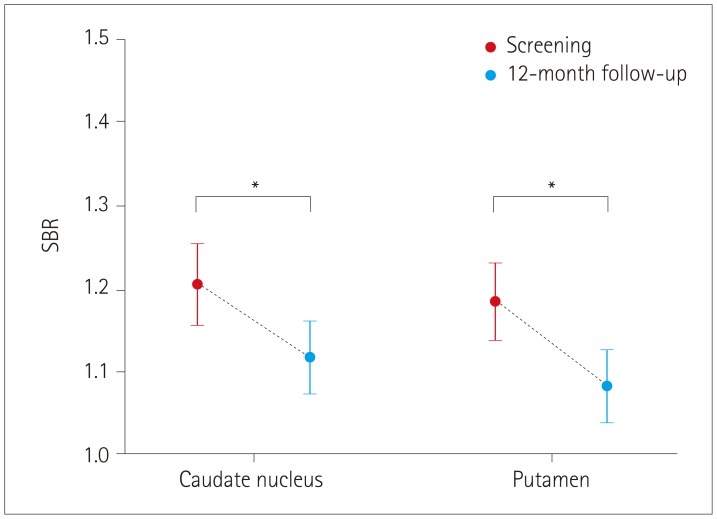
We aimed to determine the association between the annual changes in dopamine transporter (DAT) availability as measured by 123I-ioflupane (123I-FP-CIT) single-photon-emission computed tomography and single-nucleotide polymorphisms (SNPs) known to be risk factors in Parkinson's disease (PD).
METHODS
In total, 150 PD patients were included from the Parkinson's Progression Markers Initiative database. Specific SNPs that are associated with PD were selected for genotyping. SNPs that were not in Hardy-Weinberg equilibrium or whose minor allele frequency was less than 0.05 were excluded. Twenty-three SNPs met the inclusion criteria for this study. The Kruskal-Wallis test was used to compare annual percentage changes in DAT availability for three subgroups of SNP.
RESULTS
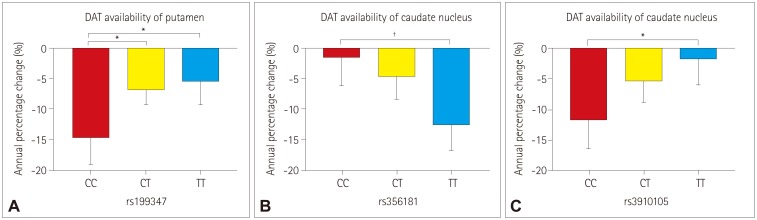
None of the 23 SNPs exerted a statistically significant effect (p < 0.0022) on the decline of DAT availability in PD patients. However, we observed trends of association (p < 0.05) between three SNPs of two genes with the annual percentage change in DAT availability: 1) rs199347 on the putamen (p=0.0138), 2) rs356181 on the caudate nucleus (p=0.0105), and 3) rs3910105 on the caudate nucleus (p=0.0374). A post-hoc analysis revealed that DAT availability was reduced the most for 1) the putamen in the CC genotype of rs199347 (vs. CT, p=0.0199; vs. TT, p=0.0164), 2) the caudate nucleus in the TT genotype of rs356181 (vs. CC, p=0.0081), and 3) the caudate nucleus in the CC genotype of rs3910105 (vs. TT, p=0.0317).
CONCLUSIONS
Significant trends in the associations between three SNPs and decline of DAT availability in PD patients have been discovered.
Keyword
MeSH Terms
Figure
Reference
-
1. Lotharius J, Brundin P. Pathogenesis of Parkinson's disease: dopamine, vesicles and alpha-synuclein. Nat Rev Neurosci. 2002; 3:932–942. PMID: 12461550.2. Thomas AJ, Attems J, Colloby SJ, O'Brien JT, McKeith I, Walker R, et al. Autopsy validation of 123I-FP-CIT dopaminergic neuroimaging for the diagnosis of DLB. Neurology. 2017; 88:276–283. PMID: 27940650.3. Park E. A new era of clinical dopamine transporter imaging using 123I-FP-CIT. J Nucl Med Technol. 2012; 40:222–228. PMID: 23160562.4. Djang DS, Janssen MJ, Bohnen N, Booij J, Henderson TA, Herholz K, et al. SNM practice guideline for dopamine transporter imaging with 123I-ioflupane SPECT 1.0. J Nucl Med. 2012; 53:154–163. PMID: 22159160.5. Suwijn SR, van Boheemen CJ, de Haan RJ, Tissingh G, Booij J, de Bie RM. The diagnostic accuracy of dopamine transporter SPECT imaging to detect nigrostriatal cell loss in patients with Parkinson's disease or clinically uncertain parkinsonism: a systematic review. EJNMMI Res. 2015; 5:12. PMID: 25853018.
Article6. Benamer HT, Patterson J, Wyper DJ, Hadley DM, Macphee GJ, Grosset DG. Correlation of Parkinson's disease severity and duration with 123I-FP-CIT SPECT striatal uptake. Mov Disord. 2000; 15:692–698. PMID: 10928580.7. Kägi G, Bhatia KP, Tolosa E. The role of DAT-SPECT in movement disorders. J Neurol Neurosurg Psychiatry. 2010; 81:5–12. PMID: 20019219.8. Billingsley KJ, Bandres-Ciga S, Saez-Atienzar S, Singleton AB. Genetic risk factors in Parkinson's disease. Cell Tissue Res. 2018; 373:9–20. PMID: 29536161.
Article9. McNeill A, Wu RM, Tzen KY, Aguiar PC, Arbelo JM, Barone P, et al. Dopaminergic neuronal imaging in genetic Parkinson's disease: insights into pathogenesis. PLoS One. 2013; 8:e69190. PMID: 23935950.
Article10. Huertas I, Jesús S, García-Gómez FJ, Lojo JA, Bernal-Bernal I, Bonilla-Toribio M, et al. Genetic factors influencing frontostriatal dysfunction and the development of dementia in Parkinson's disease. PLoS One. 2017; 12:e0175560. PMID: 28399184.
Article11. Parkinson Progression. The parkinson progression marker initiative (PPMI). Prog Neurobiol. 2011; 95:629–635. PMID: 21930184.12. García-Gómez FJ, García-Solís D, Luis-Simón FJ, Marín-Oyaga VA, Carrillo F, Mir P, et al. Elaboration of the SPM template for the standardization of SPECT images with 123I-Ioflupane. Rev Esp Med Nucl Imagen Mol. 2013; 32:350–356. PMID: 23570700.13. Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002; 15:273–289. PMID: 11771995.
Article14. Squire LR. Encyclopedia of Neuroscience. Boston: Elsevier Academic Press;2009.15. Fareed M, Afzal M. Single nucleotide polymorphism in genome-wide association of human population: a tool for broad spectrum service. Egypt J Med Hum Genet. 2013; 14:123–134.
Article16. van de Giessen E, de Win MM, Tanck MW, van den Brink W, Baas F, Booij J. Striatal dopamine transporter availability associated with polymorphisms in the dopamine transporter gene SLC6A3. J Nucl Med. 2009; 50:45–52. PMID: 19091889.
Article17. Zhai D, Li S, Zhao Y, Lin Z. SLC6A3 is a risk factor for Parkinson's disease: a meta-analysis of sixteen years' studies. Neurosci Lett. 2014; 564:99–104. PMID: 24211691.
Article18. Nalls MA, Pankratz N, Lill CM, Do CB, Hernandez DG, Saad M, et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nat Genet. 2014; 46:989–993. PMID: 25064009.19. Murthy MN, Blauwendraat C, Guelfi S, Hardy J, Lewis PA, Trabzuni D. Increased brain expression of GPNMB is associated with genome wide significant risk for Parkinson's disease on chromosome 7p15.3. Neurogenetics. 2017; 18:121–133. PMID: 28391543.
Article20. Neal ML, Boyle AM, Budge KM, Safadi FF, Richardson JR. The glycoprotein GPNMB attenuates astrocyte inflammatory responses through the CD44 receptor. J Neuroinflammation. 2018; 15:73. PMID: 29519253.
Article21. Wang Q, Liu Y, Zhou J. Neuroinflammation in Parkinson's disease and its potential as therapeutic target. Transl Neurodegener. 2015; 4:19. PMID: 26464797.
Article22. Wu HC, Chen CM, Chen YC, Fung HC, Chang KH, Wu YR. DLG2, but not TMEM229B, GPNMB, and ITGA8 polymorphism, is associated with Parkinson's disease in a Taiwanese population. Neurobiol Aging. 2018; 64:158.e1–158.e6.
Article23. Foo JN, Tan LC, Irwan ID, Au WL, Low HQ, Prakash KM, et al. Genome-wide association study of Parkinson's disease in East Asians. Hum Mol Genet. 2017; 26:226–232. PMID: 28011712.
Article24. Mata IF, Shi M, Agarwal P, Chung KA, Edwards KL, Factor SA, et al. SNCA variant associated with Parkinson disease and plasma alpha-synuclein level. Arch Neurol. 2010; 67:1350–1356. PMID: 21060011.
Article25. Kang JH, Mollenhauer B, Coffey CS, Toledo JB, Weintraub D, Galasko DR, et al. CSF biomarkers associated with disease heterogeneity in early Parkinson's disease: the Parkinson's Progression Markers Initiative study. Acta Neuropathol. 2016; 131:935–949. PMID: 27021906.
Article26. Xu W, Tan L, Yu JT. Link between the SNCA gene and parkinsonism. Neurobiol Aging. 2015; 36:1505–1518. PMID: 25554495.27. Tagliafierro L, Chiba-Falek O. Up-regulation of SNCA gene expression: implications to synucleinopathies. Neurogenetics. 2016; 17:145–157. PMID: 26948950.
Article28. Miranda-Morales E, Meier K, Sandoval-Carrillo A, Salas-Pacheco J, Vázquez-Cárdenas P, Arias-Carrión O. Implications of DNA methylation in Parkinson's disease. Front Mol Neurosci. 2017; 10:225. PMID: 28769760.
Article29. Guhathakurta S, Evangelista BA, Ghosh S, Basu S, Kim YS. Hypomethylation of intron1 of α-synuclein gene does not correlate with Parkinson's disease. Mol Brain. 2017; 10:6. PMID: 28173842.
Article30. Hernandez DM. Genetic variation and DNA methylation in the context of neurological disease [dissertation]. London: University College London;2016.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Practical Approach for the Clinical Use of Dopamine Transporter Imaging
- Classic Studies on the Interaction of Cocaine and the Dopamine Transporter
- Dopamine Receptors and Agonists in Parkinson's Disease
- Study of dopamine transporter in the rat 6-hydroxydopamine Parkinson's disease model
- Heterogeneous Patterns of Striatal Dopamine Loss in Patients with Young- versus Old-Onset Parkinson's Disease: Impact on Clinical Features