Ann Lab Med.
2019 Sep;39(5):478-487. 10.3343/alm.2019.39.5.478.
Comparison of cobas EGFR Mutation Test v2 and PANAMutyper-R-EGFR for Detection and Semi-Quantification of Epidermal Growth Factor Receptor Mutations in Plasma and Pleural Effusion Supernatant
- Affiliations
-
- 1Department of Family Medicine, School of Medicine, Wonkwang University, Iksan, Korea.
- 2Department of Internal Medicine, School of Medicine, Wonkwang University, Iksan, Korea.
- 3Department of Pathology, School of Medicine, Wonkwang University, Iksan, Korea.
- 4Department of Laboratory Medicine and Institute of Wonkwang Medical Science, School of Medicine, Wonkwang University, Iksan, Korea. emailds@hanmail.net
- 5Department of Oriental Medical Ophthalmology & Otolaryngology & Dermatology, College of Oriental Medicine, Wonkwang University, Iksan, Korea.
- 6Wonkwang Institute of Clinical Medicine, Wonkwang University Hospital, Iksan, Korea.
- KMID: 2450971
- DOI: http://doi.org/10.3343/alm.2019.39.5.478
Abstract
- BACKGROUND
Plasma epidermal growth factor receptor (EGFR) mutation tests are less invasive than tissue EGFR mutation tests. We determined which of two kits is more efficient: cobas EGFR Mutation test v2 (cobasv2; Roche Molecular Systems, Pleasanton, CA, USA) or PANAMutyper-R-EGFR (Mutyper; Panagene, Daejeon, Korea). We also evaluated whether pleural effusion supernatant (PE-SUP) samples are assayable, similar to plasma samples, using these two kits.
METHODS
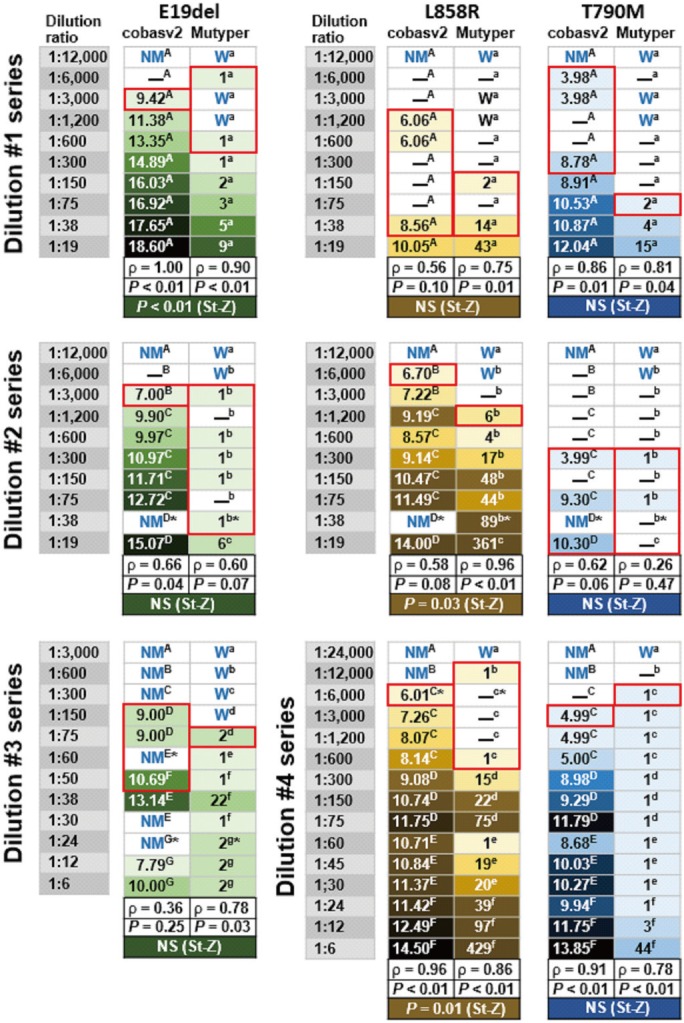
We analyzed 156 plasma and PE-SUP samples (31 paired samples) from 116 individuals. We compared the kits in terms of accuracy, assessed genotype concordance (weighted κ with 95% confidence intervals), and calculated Spearman's rho between semi-quantitatively measured EGFR-mutant levels (SQIs) measured by each kit. We also compared sensitivity using 47 EGFR-mutant harboring samples divided into more-dilute and less-dilute samples (dilution ratio: ≥ or <1:1,000).
RESULTS
cobasv2 tended to have higher accuracy than Mutyper (73% vs 69%, P=0.53), and PE-SUP samples had significantly higher accuracy than plasma samples (97% vs 55-71%) for both kits. Genotype concordance was 98% (κ=0.92, 0.88-0.96). SQIs showed strong positive correlations (P<0.0001). In less-dilute samples, accuracy and sensitivity did not differ significantly between kits. In more-dilute samples, cobasv2 tended to have higher sensitivity than Mutyper (43% vs 20%, P=0.07).
CONCLUSIONS
The kits have similar performance in terms of EGFR mutation detection and semi-quantification in plasma and PE-SUP samples. cobasv2 tends to outperform Mutyper in detecting less-abundant EGFR-mutants. PE-SUP samples are assayable using either kit.
Keyword
MeSH Terms
Figure
Reference
-
1. Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007; 7:169–181. PMID: 17318210.2. Lee CK, Brown C, Gralla RJ, Hirsh V, Thongprasert S, Tsai CM, et al. Impact of EGFR inhibitor in non-small cell lung cancer on progression-free and overall survival: a meta-analysis. J Natl Cancer Inst. 2013; 105:595–605. PMID: 23594426.3. Yu HA, Arcila ME, Rekhtman N, Sima CS, Zakowski MF, Pao W, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013; 19:2240–2247. PMID: 23470965.4. Tan DS, Yom SS, Tsao MS, Pass HI, Kelly K, Peled N, et al. The International Association for the Study of Lung Cancer consensus statement on optimizing management of EGFR mutation-positive non-small cell lung cancer: status in 2016. J Thorac Oncol. 2016; 11:946–963. PMID: 27229180.5. Mok TS, Wu YL, Ahn MJ, Garassino MC, Kim HR, Ramalingam SS, et al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med. 2017; 376:629–640. PMID: 27959700.6. Tan CS, Kumarakulasinghe NB, Huang YQ, Ang YLE, Choo JR, Goh BC, et al. Third generation EGFR TKI's: current data and future directions. Mol Cancer. 2018; 17:29. PMID: 29455654.7. Lindeman NI, Cagle PT, Aisner DL, Arcila ME, Beasley MB, Bernicker EH, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch Pathol Lab Med. 2018; 142:321–346. PMID: 29355391.8. Marchetti A, Palma JF, Felicioni L, De Pas TM, Chiari R, Del Grammastro M, et al. Early prediction of response to tyrosine kinase inhibitors by quantification of EGFR mutations in plasma of NSCLC patients. J Thorac Oncol. 2015; 10:1437–1443. PMID: 26295376.9. Tseng JS, Yang TY, Tsai CR, Chen KC, Hsu KH, Tsai MH, et al. Dynamic plasma EGFR mutation status as a predictor of EGFR-TKI efficacy in patients with EGFR-mutant lung adenocarcinoma. J Thorac Oncol. 2015; 10:603–610. PMID: 25514801.10. Singh AP, Li S, Cheng H. Circulating DNA in EGFR-mutated lung cancer. Ann Transl Med. 2017; 5:379. PMID: 29057239.11. Vendrell JA, Mau-Them FT, Béganton B, Godreuil S, Coopman P, Solassol J. Circulating cell free tumor DNA detection as a routine tool for lung cancer patient management. Int J Mol Sci. 2017; 18:E264. PMID: 28146051.12. Kim CG, Shim HS, Hong MH, Cha YJ, Heo SJ, Park HS, et al. Detection of activating and acquired resistant mutation in plasma from EGFR-mutated NSCLC patients by peptide nucleic acid (PNA) clamping-assisted fluorescence melting curve analysis. Oncotarget. 2017; 8:65111–65122. PMID: 29029416.13. Barlesi F, Mazieres J, Merlio JP, Debieuvre D, Mosser J, Lena H, et al. Routine molecular profiling of patients with advanced non-small-cell lung cancer: results of a 1-year nationwide programme of the French Cooperative Thoracic InterGroup (IFCT). Lancet. 2016; 387:1415–1426. PMID: 26777916.14. Karlovich C, Goldman JW, Sun JM, Mann E, Sequist LV, Konopa K, et al. Assessment of EGFR mutation status in matched plasma and tumor tissue of NSCLC patients from a Phase I study of rociletinib (CO-1686). Clin Cancer Res. 2016; 22:2386–2395. PMID: 26747242.15. Mok T, Wu YL, Lee JS, Yu CJ, Sriuranpong V, Sandoval-Tan J, et al. Detection and dynamic changes of EGFR mutations from circulating tumor DNA as a predictor of survival outcomes in NSCLC patients treated with first-line intercalated erlotinib and chemotherapy. Clin Cancer Res. 2015; 21:3196–3203. PMID: 25829397.16. Piotrowska Z, Niederst MJ, Karlovich CA, Wakelee HA, Neal JW, Mino-Kenudson M, et al. Heterogeneity underlies the emergence of EGFR T790 wild-type clones following treatment of T790M-positive cancers with a third-generation EGFR inhibitor. Cancer Discov. 2015; 5:713–722. PMID: 25934077.17. Alegre E, Fusco JP, Restituto P, Salas-Benito D, Rodríguez-Ruiz ME, Andueza MP, et al. Total and mutated EGFR quantification in cell-free DNA from non-small cell lung cancer patients detects tumor heterogeneity and presents prognostic value. Tumour Biol. 2016; 37:13687–13694. PMID: 27473086.18. Diedenhofen B, Musch J. Cocor: a comprehensive solution for the statistical comparison of correlations. PLoS One. 2015; 10:e0121945. PMID: 25835001.19. Popper HH. Progression and metastasis of lung cancer. Cancer Metastasis Rev. 2016; 35:75–91. PMID: 27018053.20. Liu X, Lu Y, Zhu G, Lei Y, Zheng L, Qin H, et al. The diagnostic accuracy of pleural effusion and plasma samples versus tumour tissue for detection of EGFR mutation in patients with advanced non-small cell lung cancer: comparison of methodologies. J Clin Pathol. 2013; 66:1065–1069. PMID: 23888061.21. Yeo CD, Kim JW, Kim KH, Ha JH, Rhee CK, Kim SJ, et al. Detection and comparison of EGFR mutations in matched tumor tissues, cell blocks, pleural effusions, and sera from patients with NSCLC with malignant pleural effusion, by PNA clamping and direct sequencing. Lung Cancer. 2013; 81:207–212. PMID: 23726527.22. Schrader C, Schielke A, Ellerbroek L, Johne R. PCR inhibitors-occurrence, properties and removal. J Appl Microbiol. 2012; 113:1014–1026. PMID: 22747964.23. Hotta K, Zhu CB, Phomsuwansiri P, Ishikawa J, Mizuno S, Hatsu M, et al. PCR inhibition assay for DNA-targeted antibiotics. J Antibiot (Tokyo). 1995; 48:1267–1272. PMID: 8557567.24. Sidstedt M, Hedman J, Romsos EL, Waitara L, Wadsö L, Steffen CR, et al. Inhibition mechanisms of hemoglobin, immunoglobulin G, and whole blood in digital and real-time PCR. Anal Bioanal Chem. 2018; 410:2569–2583. PMID: 29504082.25. Field Safety Corrective Action, Potential inhibition of plasma samples with the cobas EGFR Mutation test, v2 CE-IVD when used in conjunction with the cobas cfDNA Sample Preparation Kit, MDFA News. Updated on May 2016. http://www.mfds.go.kr/eng/brd/m_52/list.do?multi_itm_seq=0&srchTp=0&srchWord=EGFR+Mutation.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Landscape of EGFR mutations in lung adenocarcinoma: a single institute experience with comparison of PANAMutyper testing and targeted next-generation sequencing
- Mutations of the Epidermal Growth Factor Receptor Gene in Triple-Negative Breast Cancer
- EGFR Mutation Status in Lung Adenocarcinoma-Associated Malignant Pleural Effusion and Efficacy of EGFR Tyrosine Kinase Inhibitors
- Does the efficacy of epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor differ according to the type of EGFR mutation in non-small cell lung cancer?
- A Case of Patient with Lung Adenocarcinoma with Double Rare EGFR Mutation of G719C and L861Q