Ann Lab Med.
2019 Nov;39(6):515-523. 10.3343/alm.2019.39.6.515.
Korean Society for Genetic Diagnostics Guidelines for Validation of Next-Generation Sequencing-Based Somatic Variant Detection in Hematologic Malignancies
- Affiliations
-
- 1Department of Laboratory Medicine & Genetics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea. kimjw@skku.edu
- 2Department of Laboratory Medicine, Korea Cancer Center Hospital, Seoul, Korea.
- 3Department of Laboratory Medicine, Yonsei University College of Medicine, Seoul, Korea.
- KMID: 2450945
- DOI: http://doi.org/10.3343/alm.2019.39.6.515
Abstract
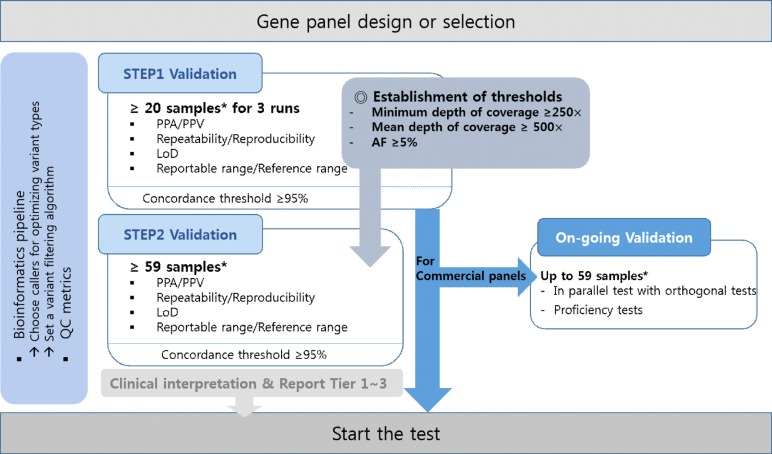
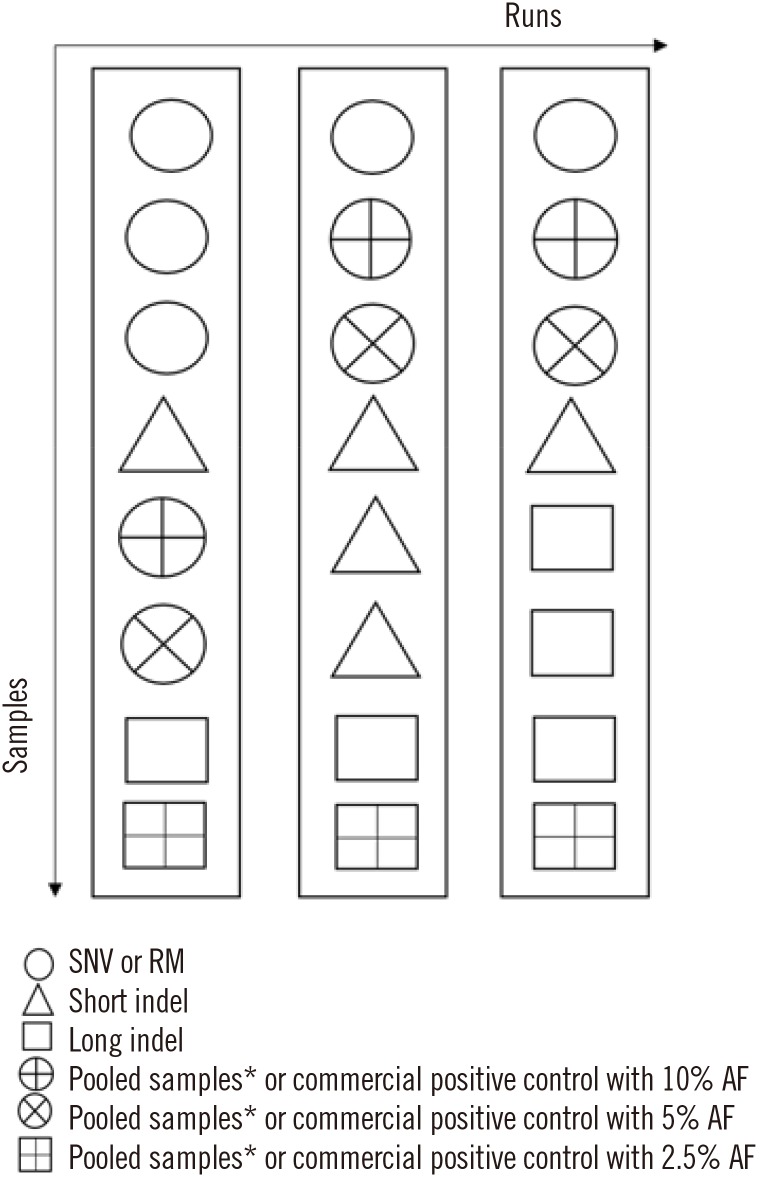
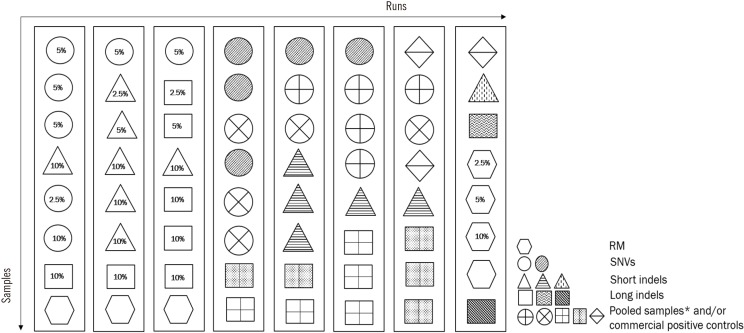
- Next-generation sequencing (NGS) is currently used in the clinical setting for targeted therapies and diagnosis of hematologic malignancies. Accurate detection of somatic variants is challenging because of tumor purity, heterogeneity, and the complexity of genetic alterations, with various issues ranging from high detection design to test implementation. This article presents guidelines developed through consensus among a panel of experts from the Korean Society for Genetic Diagnostics. They are based on experiences with the validation processes of NGS-based somatic panels for hematologic malignancies, with reference to previous international recommendations. These guidelines describe basic parameters with emphasis on the design of a validation protocol for NGS-based somatic panels to be used in practice. In addition, they suggest thresholds of key metrics, including minimum coverage, mean coverage with uniformity index, and minimum variant allele frequency, for the initial diagnosis of hematologic malignancies.
Keyword
Figure
Cited by 2 articles
-
Application of Next Generation Sequencing in Laboratory Medicine
Yiming Zhong, Feng Xu, Jinhua Wu, Jeffrey Schubert, Marilyn M. Li
Ann Lab Med. 2021;41(1):25-43. doi: 10.3343/alm.2021.41.1.25.Measurable Residual Disease Testing Using Next-Generation Sequencing in Acute Myeloid Leukemia
Seon Young Kim, Hee Jin Huh
Ann Lab Med. 2023;43(4):323-324. doi: 10.3343/alm.2023.43.4.323.
Reference
-
1. Watson IR, Takahashi K, Futreal PA, Chin L. Emerging patterns of somatic mutations in cancer. Nat Rev Genet. 2013; 14:703–718. PMID: 24022702.2. Coombs CC, Tallman MS, Levine RL. Molecular therapy for acute myeloid leukaemia. Nat Rev Clin Oncol. 2016; 13:305–318. PMID: 26620272.3. Galanina N, Bejar R, Choi M, Goodman A, Wieduwilt M, Mulroney C, et al. Comprehensive genomic profiling reveals diverse but actionable molecular portfolios across hematologic malignancies: implications for next generation clinical trials. Cancers (Basel). 2018; 11.4. Vainchenker W, Kralovics R. Genetic basis and molecular pathophysiology of classical myeloproliferative neoplasms. Blood. 2017; 129:667–679. PMID: 28028029.5. Grinfeld J, Nangalia J, Green AR. Molecular determinants of pathogenesis and clinical phenotype in myeloproliferative neoplasms. Haematologica. 2017; 102:7–17. PMID: 27909216.6. Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012; 481:506–510. PMID: 22237025.7. Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016; 127:2391–2405. PMID: 27069254.8. Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012; 366:883–892. PMID: 22397650.9. Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016; 374:2209–2221. PMID: 27276561.10. Goodman AM, Choi M, Wieduwilt M, Mulroney C, Costello C, Frampton G, et al. Next generation sequencing reveals potentially actionable alterations in the majority of patients with lymphoid malignancies. JCO Precis Oncol. 2017; 1:1–13.11. Taylor J, Xiao W, Abdel-Wahab O. Diagnosis and classification of hematologic malignancies on the basis of genetics. Blood. 2017; 130:410–423. PMID: 28600336.12. Ballester LY, Luthra R, Kanagal-Shamanna R, Singh RR. Advances in clinical next-generation sequencing: target enrichment and sequencing technologies. Expert Rev Mol Diagn. 2016; 16:357–372. PMID: 26680590.13. Kanagal-Shamanna R, Singh RR, Routbort MJ, Patel KP, Medeiros LJ, Luthra R. Principles of analytical validation of next-generation sequencing based mutational analysis for hematologic neoplasms in a CLIA-certified laboratory. Expert Rev Mol Diagn. 2016; 16:461–472. PMID: 26765348.14. Yohe S, Thyagarajan B. Review of clinical next-generation sequencing. Arch Pathol Lab Med. 2017; 141:1544–1557. PMID: 28782984.15. Swerdlow SH, Campo E, et al. WHO classification of tumours of haematopoietic and lymphoid tissues. revised 4th ed. Lyon: International Agency for Research on Cancer;2017. p. 16–27.16. Jennings LJ, Arcila ME, Corless C, Kamel-Reid S, Lubin IM, Pfeifer J, et al. Guidelines for validation of next-generation sequencing-based oncology panels: a joint consensus recommendation of the association for molecular pathology and college of American Pathologists. J Mol Diagn. 2017; 19:341–365. PMID: 28341590.17. Gargis AS, Kalman L, Berry MW, Bick DP, Dimmock DP, Hambuch T, et al. Assuring the quality of next-generation sequencing in clinical laboratory practice. Nat Biotechnol. 2012; 30:1033–1036. PMID: 23138292.18. Matthijs G, Souche E, Alders M, Corveleyn A, Eck S, Feenstra I, et al. Guidelines for diagnostic next-generation sequencing. Eur J Hum Genet. 2016; 24:1515.19. Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017; 129:424–447. PMID: 27895058.20. Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: A joint consensus recommendation of the association for molecular pathology, American Society of Clinical Oncology, and College of American pathologists. J Mol Diagn. 2017; 19:4–23. PMID: 27993330.21. Ellard S, Lindsay H, Camm N, Watson C, Abbs S, Mattocks C, et al. Practice guidelines for targeted next generation sequencing analysis and interpretation. Association for clinical genetic science (ACGS). https://www.acgs.uk.com/media/10789/bpg_for_targeted_next_generation_sequencing_-_approved_dec_2015.pdf.22. Rehm HL, Bale SJ, Bayrak-Toydemir P, Berg JS, Brown KK, Deignan JL, et al. ACMG clinical laboratory standards for next-generation sequencing. Genet Med. 2013; 15:733–747. PMID: 23887774.23. Ben Lassoued A, Nivaggioni V, Gabert J. Minimal residual disease testing in hematologic malignancies and solid cancer. Expert Rev Mol Diagn. 2014; 14:699–712. PMID: 24938122.24. Cuomo AW, Zucker HA, Dreslin S. Oncology-molecular and cellular tumor markers “Next Generation” sequencing (NGS) guidelines for somatic genetic variant detection. Updated on Jan 2018. https://www.wadsworth.org/regulatory/clep/clinical-labs/obtain-permit/test-approval.25. Beck TF, Mullikin JC, Biesecker LG. NISC Comparative Sequencing Program. Systematic evaluation of Sanger validation of next-generation sequencing variants. Clin Chem. 2016; 62:647–654. PMID: 26847218.26. Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013; 43:11.10.1–11.10.33. PMID: 25431634.27. Shin HT, Choi YL, Yun JW, Kim NKD, Kim SY, Jeon HJ, et al. Prevalence and detection of low-allele-fraction variants in clinical cancer samples. Nat Commun. 2017; 8:1377. PMID: 29123093.28. Wala JA, Bandopadhayay P, Greenwald NF, O'Rourke R, Sharpe T, Stewart C, et al. SvABA: genome-wide detection of structural variants and indels by local assembly. Genome Res. 2018; 28:581–591. PMID: 29535149.29. Scheinin I, Sie D, Bengtsson H, van de Wiel MA, Olshen AB, van Thuijl HF, et al. DNA copy number analysis of fresh and formalin-fixed specimens by shallow whole-genome sequencing with identification and exclusion of problematic regions in the genome assembly. Genome Res. 2014; 24:2022–2032. PMID: 25236618.30. Levin JZ, Berger MF, Adiconis X, Rogov P, Melnikov A, Fennell T, et al. Targeted next-generation sequencing of a cancer transcriptome enhances detection of sequence variants and novel fusion transcripts. Genome Biol. 2009; 10:R115. PMID: 19835606.31. Kumar S, Razzaq SK, Vo AD, Gautam M, Li H. Identifying fusion transcripts using next generation sequencing. Wiley Interdiscip Rev RNA. 2016; 7:811–823. PMID: 27485475.32. Liu S, Tsai WH, Ding Y, Chen R, Fang Z, Huo Z, et al. Comprehensive evaluation of fusion transcript detection algorithms and a meta-caller to combine top performing methods in paired-end RNA-seq data. Nucleic Acids Res. 2016; 44:e47. PMID: 26582927.33. Zeng X, Lin W, Guo M, Zou Q. A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput Biol. 2017; 13:e1005420. PMID: 28594838.34. Liu S, Tsai WH, Ding Y, Chen R, Fang Z, Huo Z, et al. Comprehensive evaluation of fusion transcript detection algorithms and a meta-caller to combine top performing methods in paired-end RNA-seq data. Nucleic Acids Res. 2016; 44:e47. PMID: 26582927.35. Singh RR, Luthra R, Routbort MJ, Patel KP, Medeiros LJ. Implementation of next generation sequencing in clinical molecular diagnostic laboratories: advantages, challenges and potential. Expert Rev Precis Med Drug Dev. 2016; 1:109–120.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Status of Next-Generation Sequencing-Based Genetic Diagnosis in Hematologic Malignancies in Korea (2017-2018)
- Report of the Korean Association of External Quality Assessment Service on Next-Generation Sequencing Analysis for Somatic Variants (2018–2020)
- Guidelines for Genetic Counseling and Reporting of Cancer Genetic Test Results: Genetic Test for Hereditary Cancer-predisposing Syndrome Using Next-generation Sequencing
- Detection of ASXL1 Codon 646 Variant Using Amplicon-Based Next-Generation Sequencing
- Detection of Mosaic Sequence Variants Associated with Human Genetic Diseases