Ann Lab Med.
2018 Sep;38(5):466-472. 10.3343/alm.2018.38.5.466.
The Role of the Signal-to-Cutoff Ratio in Automated Anti-HCV Chemiluminescent Immunoassays by Referring to the Nucleic Acid Amplification Test and the Recombinant Immunoblot Assay
- Affiliations
-
- 1Department of Laboratory Medicine, Seoul National University Bundang Hospital, Seongnam, Korea. aeiea@snu.ac.kr
- 2Department of Laboratory Medicine, Chonbuk National University College of Medicine, Jeonju, Korea.
- 3Department of Laboratory Medicine, Seoul National University College of Medicine, Seoul, Korea.
- KMID: 2434737
- DOI: http://doi.org/10.3343/alm.2018.38.5.466
Abstract
- BACKGROUND
Following discontinuation of the recombinant immunoblot assay (RIBA), the only available supplementary test for the detection of hepatitis C virus (HCV) is the nucleic acid amplification test (NAAT). However, the NAAT does not adequately detect past HCV. Consequently, it is hard to distinguish between past HCV infection and biological false positivity with an anti-HCV result alone. We assessed the diagnostic performance of two immunoassays: the ARCHITECT anti-HCV chemiluminescent microparticle immunoassay (CMIA; Abbott Diagnostics, Wiesbaden, Germany) and the Access HCV Ab PLUS chemiluminescent immunoassay (CIA; Bio-Rad, Marnes-la-Coquette, France). We also explored an optimized algorithm to determine the anti-HCV results.
METHODS
We tested 126,919 patients and 44,556 individuals who underwent a medical checkup. RIBA and NAAT were conducted for samples that tested anti-HCV-positive using CMIA and CIA. We assessed the optimal signal-to-cutoff (S/CO) ratio in HCV-positive samples.
RESULTS
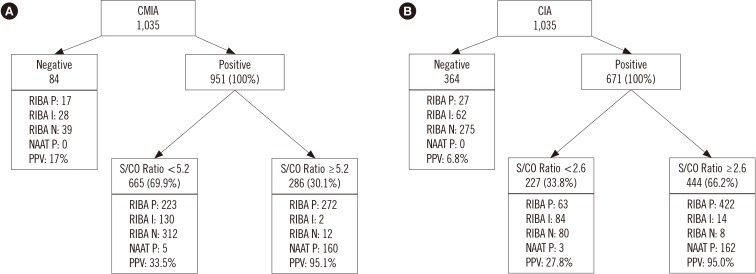
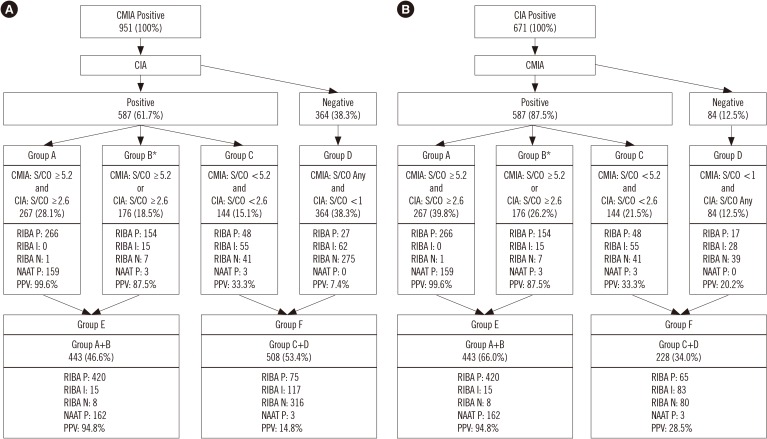
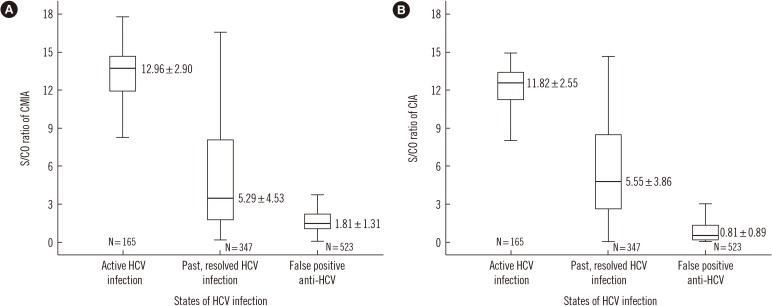
In total, 1,035 blood samples tested anti-HCV-positive. Of these, RIBA was positive in 512, indeterminate in 160, and negative in 363 samples. One hundred sixty-five samples were NAAT-positive. Diagnostic sensitivity and positive predictive value (PPV) were 96.7% and 52.1%, respectively, for CMIA, and 94.7% and 72.3%, respectively, for CIA. The optimal S/CO ratio was 5.2 for CMIA and 2.6 for CIA at 95% PPV. In total, 286 samples tested positive in CMIA and 444 in CIA, while 443 samples tested positive in both assays.
CONCLUSIONS
It is hard to determine anti-HCV positivity based on the S/CO ratio alone. However, this study elucidated the role of the S/CO ratio by using the NAAT and RIBA.
Keyword
Figure
Cited by 2 articles
-
The impact of nucleic acid testing as a blood donor screening method in transfusion-associated hepatitis C among children with bleeding disorders in Indonesia: a single-center experience
Novie Amelia Chozie, Melati Arum Satiti, Damayanti Rusli Sjarif, Hanifah Oswari, Ni Ken Ritchie
Blood Res. 2022;57(2):129-134. doi: 10.5045/br.2022.2021219.Cross-Reactivity of Disease-Specific Antibody Assays for the Detection of Current Infections: With Potentially Interfering Substances of Other Infections
Dae Ho Gong, Seung Hyeon Kim, Hee-Jeong Kim, Anna Lee, Mi-Soon Han
Lab Med Qual Assur. 2022;44(1):40-47. doi: 10.15263/jlmqa.2022.44.1.40.
Reference
-
1. Ghany MG, Strader DB, Thomas DL, Seeff LB. American Association for the Study of Liver Diseases. Diagnosis, management, and treatment of hepatitis C: an update. Hepatology. 2009; 49:1335–1374. PMID: 19330875.2. Korean Association for the Study of the Liver. Chronic Hepatitis C Clinical Practice Guideline [guidelines]. Updated on Apr, 2018. http://www.kasl.org/bbs/index.html?code=guide&category.3. Pawlotsky JM. Use and interpretation of virological tests for hepatitis C. Hepatology. 2002; 36(Suppl 1):S65–S73. PMID: 12407578.4. World Health Organization. Guidelines for the Screening Care and Treatment of Persons with Chronic Hepatitis C Infection: Updated Version [guidelines]. Updated on Apr, 2018. http://www.who.int/hepatitis/publications/hepatitis-c-guidelines-2016/en/.5. Pereira FM, Zarife MA, Reis EA, G Reis M. Indeterminate RIBA results were associated with the absence of hepatitis C virus RNA (HCV-RNA) in blood donors. Rev Soc Bras Med Trop. 2014; 47:12–17. PMID: 24603731.6. Berger A, Rabenau H, Allwinn R, Doerr HW. Evaluation of the new ARCHITECT anti-HCV screening test under routine laboratory conditions. J Clin Virol. 2008; 43:158–161. PMID: 18635393.7. Alter MJ, Kruszon-Moran D, Nainan OV, McQuillan GM, Gao F, Moyer LA, et al. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N Engl J Med. 1999; 341:556–562. PMID: 10451460.8. Conry-Cantilena C, VanRaden M, Gibble J, Melpolder J, Shakil AO, Viladomiu L, et al. Routes of infection, viremia, and liver disease in blood donors found to have hepatitis C virus infection. N Engl J Med. 1996; 334:1691–1696. PMID: 8637513.9. Goetz AM, Ndimbie OK, Wagener MM, Muder RR. Prevalence of hepatitis C infection in health care workers affiliated with a liver transplant center. Transplantation. 1995; 59:990–994. PMID: 7535961.10. Gunn RA, Murray PJ, Ackers ML, Hardison WG, Margolis HS. Screening for chronic hepatitis B and C virus infections in an urban sexually transmitted disease clinic: rationale for integrating services. Sex Transm Dis. 2001; 28:166–170. PMID: 11289199.11. Hyams KC, Riddle J, Rubertone M, Trump D, Alter MJ, Cruess DF, et al. Prevalence and incidence of hepatitis C virus infection in the US military: a seroepidemiologic survey of 21,000 troops. Am J Epidemiol. 2001; 153:764–770. PMID: 11296148.12. Kleinman S, Alter H, Busch M, Holland P, Tegtmeier G, Nelles M, et al. Increased detection of hepatitis C virus (HCV)-infected blood donors by a multiple-antigen HCV enzyme immunoassay. Transfusion. 1992; 32:805–813. PMID: 1471243.13. Montecalvo MA, Lee MS, DePalma H, Wynn PS, Lowenfels AB, Jorde U, et al. Seroprevalence of human immunodeficiency virus-1, hepatitis B virus, and hepatitis C virus in patients having major surgery. Infect Control Hosp Epidemiol. 1995; 16:627–632. PMID: 8601681.14. Roome AJ, Hadler JL, Thomas AL, Migicovsky B, Roth R, Boraz M, et al. Hepatitis C virus infection among firefighters, emergency medical technicians, and paramedics--selected locations, United States, 1991-2000. MMWR Morb Mortal Wkly Rep. 2000; 49:660–665. PMID: 10943253.15. Maasoumy B, Bremer B, Raupach R, Lehmann P, Manns MP, Cornberg M, et al. How to interpret borderline HCV antibody test results: a comparative study investigating four different anti-HCV assays. Viral Immunol. 2014; 27:7–13. PMID: 24494968.16. Acar A, Kemahli S, Altunay H, Kosan E, Oncul O, Gorenek L, et al. The significance of repeat testing in Turkish blood donors screened with HBV, HCV and HIV immunoassays and the importance of S/CO ratios in the interpretation of HCV/HIV screening test results and as a determinant for further confirmatory testing. Transfus Med. 2010; 20:152–159. PMID: 20059750.17. Moretti M, Pieretti B, Masucci A, Sisti D, Rocchi M, Delprete E. Role of signal-to-cutoff ratios in hepatitis C virus antibody detection. Clin Vaccine Immunol. 2012; 19:1329–1331. PMID: 22695164.18. Stramer SL, Dodd RY, Brodsky JP. The value of screening signal-to-cutoff ratios for hepatitis C virus antibody confirmation. Transfusion. 2013; 53:1497–1500. PMID: 23176156.19. Alter MJ, Kuhnert WL, Finelli L. Centers for Disease Control and Prevention. Guidelines for laboratory testing and result reporting of antibody to hepatitis C virus. Centers for Disease Control and Prevention. MMWR Recomm Rep. 2003; 52:1–13. 15quiz CE1-4.20. Centers for Disease Control and Prevention. Testing for HCV infection: an update of guidance for clinicians and laboratorians. MMWR Morb Mortal Wkly Rep. 2013; 62:362–365. PMID: 23657112.21. Lai KK, Jin M, Yuan S, Larson MF, Dominitz JA, Bankson DD. Improved reflexive testing algorithm for hepatitis C infection using signal-to-cutoff ratios of a hepatitis C virus antibody assay. Clin Chem. 2011; 57:1050–1056. PMID: 21566071.22. AASLD/IDSA HCV Guidance Panel. Hepatitis C guidance: AASLD-IDSA recommendations for testing, managing, and treating adults infected with hepatitis C virus. Hepatology. 2015; 62:932–954. PMID: 26111063.23. Vrielink H, Reesink HW, Zaaijer HL, Scholten E, Kremer LC, Cuypers HT, et al. Look-back of anti-HCV ELISA-positive, HCV-RNA PCR-negative donors and recipients of their blood products. Vox Sang. 1997; 72:67–70. PMID: 9088071.24. Vrielink H, van der Poel CL, Reesink HW, Zaaijer HL, Scholten E, Kremer LC, et al. Look-back study of infectivity of anti-HCV ELISA-positive blood components. Lancet. 1995; 345:95–96. PMID: 7815889.25. Newcombe RG. Two-sided confidence intervals for the single proportion: comparison of seven methods. Stat Med. 1998; 17:857–872. PMID: 9595616.26. Ballani N, Gupta E. Low signal-to-cutoff ratio (S/Co) in the diagnosis of hepatitis C: a diagnostic dilemma? Indian J Gastroenterol. 2015; 34:413–414. PMID: 26498021.27. Sommese L, Iannone C, Cacciatore F, De Iorio G, Napoli C. Comparison between screening and confirmatory serological assays in blood donors in a region of South Italy. J Clin Lab Anal. 2014; 28:198–203. PMID: 24478048.28. Pawlotsky JM, Lonjon I, Hezode C, Raynard B, Darthuy F, Remire J, et al. What strategy should be used for diagnosis of hepatitis C virus infection in clinical laboratories? Hepatology. 1998; 27:1700–1702. PMID: 9620345.29. Contreras AM, Ochoa-Jimenez RJ, Celis A, Mendez C, Olivares L, Rebolledo CE, et al. High antibody level: an accurate serologic marker of viremia in asymptomatic people with hepatitis C infection. Transfusion. 2010; 50:1335–1343. PMID: 20088833.30. Gong S, Schmotzer CL, Zhou L. Evaluation of quantitative real-time PCR as a hepatitis C virus supplementary test after RIBA discontinuation. J Clin Lab Anal. 2016; 30:418–423. PMID: 26499369.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Correlation between the S/CO Ratio of Anti-HCV ELISA, and the Results of the RIBA and RNA Test
- Qualitative and Quantitative Measurements of Anti-HCV Positive Blood Donor Group
- Consideration of Appropriateness of Application of Immunoblot Assay as a Reentry Test for HCV or HIV Screening Reactive Donors
- Investigation of the Association of HCV or HIV Markers in Non-Discriminated Blood Donations
- Performance of HCV and HIV-1 Nucleic Acid Amplification Test(NAT) in Korean Blood Donors