Korean J Physiol Pharmacol.
2018 Nov;22(6):617-625. 10.4196/kjpp.2018.22.6.617.
The Nedd8-activating enzyme inhibitor MLN4924 suppresses colon cancer cell growth via triggering autophagy
- Affiliations
-
- 1Department of General Surgery, 210 Hospital of Chinese People's Liberation Army, Dalian 116021, China. gaojianjun210@163.com
- 2Department of Gynecology and Obstetrics, Affiliated Zhongshan Hospital of Dalian University, Dalian 116001, China.
- KMID: 2430084
- DOI: http://doi.org/10.4196/kjpp.2018.22.6.617
Abstract
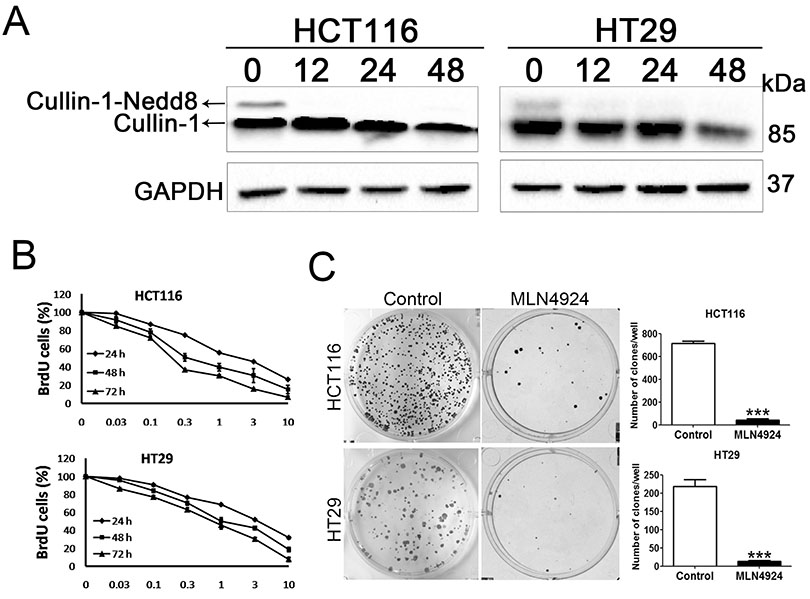
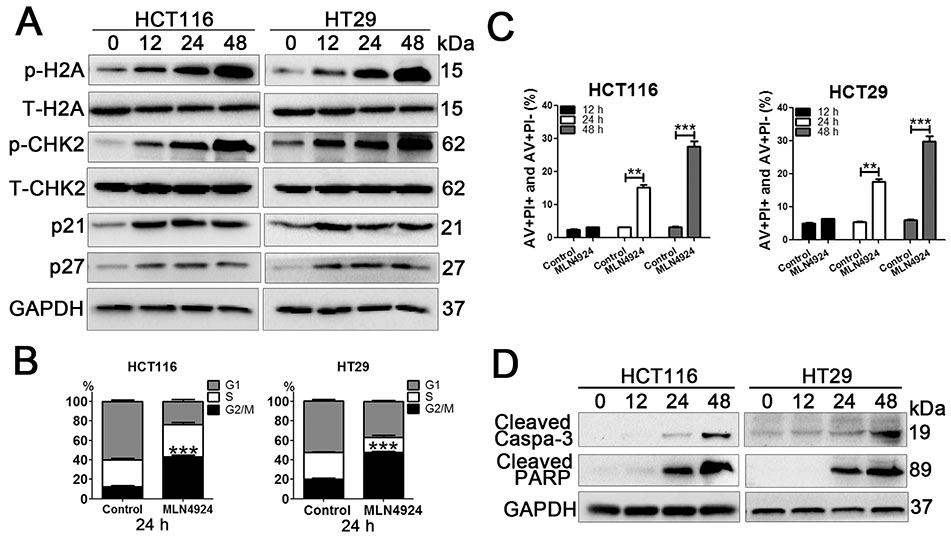
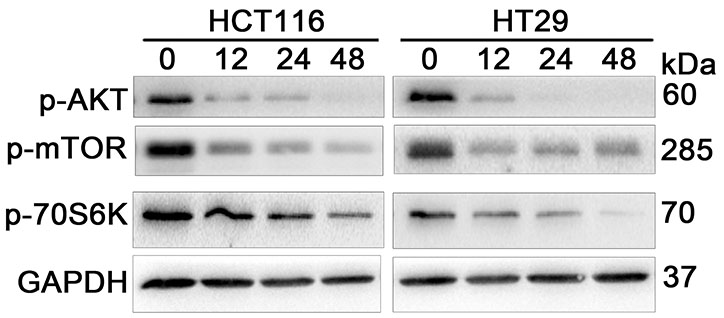
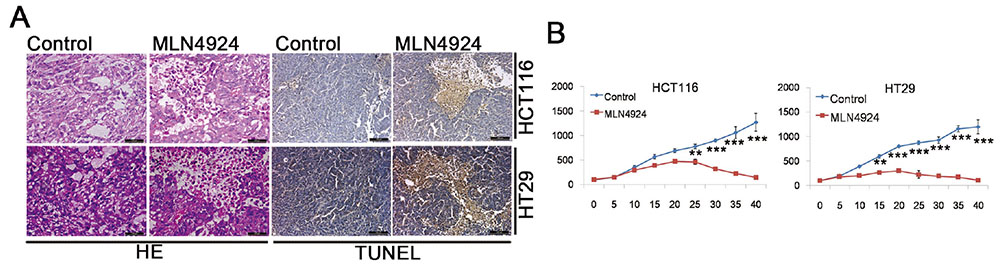
- Neddylation is a post-translational protein modification process. MLN4924 is a newly discovered pharmaceutical neddylation inhibitor that suppresses cancer growth with several cancer types. In our study, we first investigated the effect of MLN4924 on colon cancer cells (HCT116 and HT29). MLN4924 significantly inhibited the neddylation of cullin-1 and colon cancer cell growth in a time and dose-dependent manner. MLN4924 induced G2/M cell cycle arrest and apoptosis in HCT116 and HT29 cells. Moreover, MLN4924 also triggered autophagy in HCT116 and HT29 cells via suppressing the PI3K/AKT/mTOR pathway. Inhibiting autophagy by autophagy inhibitor 3-MA or ATG5 knockdown reversed the function of MLN4924 in suppressing colon cancer cell growth and cell death. Interestingly, MLN4924 suppresses colon cell growth in a xenograft model. Together, our finding revealed that blocking neddylation is an attractive colon cancer therapy strategy, and autophagy might act as a novel anti-cancer mechanism for the treatment of colon cancer by MLN4924.
Keyword
MeSH Terms
Figure
Reference
-
1. Maxwell PJ IV, Camp ER. Current controversies in the management of colon and rectal cancer. Cham: Springer International Publishing;2016.2. Petrelli F, Barni S. Correlation of progression-free and post-progression survival with overall survival in advanced colorectal cancer. Ann Oncol. 2013; 24:186–192.
Article3. Heervä E, Lavonius M, Jaakkola P, Minn H, Ristamäki R. Overall survival and metastasis resections in patients with metastatic colorectal cancer using electronic medical records. J Gastrointest Cancer. 2018; 49:245–251.
Article4. Kenific CM, Debnath J. Cellular and metabolic functions for autophagy in cancer cells. Trends Cell Biol. 2015; 25:37–45.
Article5. Ouyang L, Shi Z, Zhao S, Wang FT, Zhou TT, Liu B, Bao JK. Programmed cell death pathways in cancer: a review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012; 45:487–498.
Article6. Gil J, Pesz KA, Sąsiadek MM. May autophagy be a novel biomarker and antitumor target in colorectal cancer? Biomark Med. 2016; 10:1081–1094.
Article7. Watson IR, Irwin MS, Ohh M. NEDD8 pathways in cancer, Sine Quibus Non. Cancer Cell. 2011; 19:168–176.
Article8. Merlet J, Burger J, Gomes JE, Pintard L. Regulation of cullin-RING E3 ubiquitin-ligases by neddylation and dimerization. Cell Mol Life Sci. 2009; 66:1924–1938.
Article9. Sui Y, Liu Y, Xu G. A lysine-to-arginine mutation on NEDD8 markedly reduces the activity of cullin RING E3 ligase through the impairment of neddylation cascades. Biochem Biophys Res Commun. 2015; 461:653–658.
Article10. Xu J, Li L, Yu G, Ying W, Gao Q, Zhang W, Li X, Ding C, Jiang Y, Wei D, Duan S, Lei Q, Li P, Shi T, Qian X, Qin J, Jia L. The neddylation-cullin 2-RBX1 E3 ligase axis targets tumor suppressor RhoB for degradation in liver cancer. Mol Cell Proteomics. 2015; 14:499–509.
Article11. Wang Y, Luo Z, Pan Y, Wang W, Zhou X, Jeong LS, Chu Y, Liu J, Jia L. Targeting protein neddylation with an NEDD8-activating enzyme inhibitor MLN4924 induced apoptosis or senescence in human lymphoma cells. Cancer Biol Ther. 2015; 16:420–429.
Article12. Zhao Y, Morgan MA, Sun Y. Targeting Neddylation pathways to inactivate cullin-RING ligases for anticancer therapy. Antioxid Redox Signal. 2014; 21:2383–2400.
Article13. Nawrocki ST, Griffin P, Kelly KR, Carew JS. MLN4924: a novel firstin-class inhibitor of NEDD8-activating enzyme for cancer therapy. Expert Opin Investig Drugs. 2012; 21:1563–1573.
Article14. Lan H, Tang Z, Jin H, Sun Y. Neddylation inhibitor MLN4924 suppresses growth and migration of human gastric cancer cells. Sci Rep. 2016; 6:24218.
Article15. Zhang Y, Shi CC, Zhang HP, Li GQ, Li SS. MLN4924 suppresses neddylation and induces cell cycle arrest, senescence, and apoptosis in human osteosarcoma. Oncotarget. 2016; 7:45263–45274.
Article16. Ryabaya OO, Egorova AV, Stepanova EV. The role of autophagy in mechanisms of tumor cell death. Biol Bull Rev. 2015; 5:579–588.
Article17. Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescenttagged LC3. Autophagy. 2007; 3:452–460.
Article18. Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008; 4:151–175.19. Jiang K, Liu M, Lin G, Mao B, Cheng W, Liu H, Gal J, Zhu H, Yuan Z, Deng W, Liu Q, Gong P, Bi X, Meng S. Tumor suppressor Spred2 interaction with LC3 promotes autophagosome maturation and induces autophagy-dependent cell death. Oncotarget. 2016; 7:25652–25667.
Article20. Deng S, Tang S, Zhang S, Zhang C, Wang C, Zhou Y, Dai C, Xiao X. Furazolidone induces apoptosis through activating reactive oxygen species-dependent mitochondrial signaling pathway and suppressing PI3K/Akt signaling pathway in HepG2 cells. Food Chem Toxicol. 2015; 75:173–186.
Article21. Zheng W, Luo Z, Zhang J, Min P, Li W, Xu D, Zhang Z, Xiong P, Liang H, Liu J. Neural precursor cell expressed, developmentally downregulated 8-activating enzyme inhibitor MLN4924 sensitizes colorectal cancer cells to oxaliplatin by inducing DNA damage, G2 cell cycle arrest and apoptosis. Mol Med Rep. 2017; 15:2795–2801.
Article22. Wan J, Zhu J, Li G, Zhang Z. Radiosensitization of human colorectal cancer cells by MLN4924: an inhibitor of NEDD8-activating enzyme. Technol Cancer Res Treat. 2016; 15:527–534.23. Koskela A, Reinisalo M, Kaarniranta K. Taking a roller coaster ride with autophagy markers p62 and LC3. Acta Ophthalmol. 2016; 94:S256.
Article24. Heras-Sandoval D, Pérez-Rojas JM, Hernández-Damián J, Pedraza-Chaverri J. The role of PI3K/AKT/mTOR pathway in the modulation of autophagy and the clearance of protein aggregates in neurodegeneration. Cell Signal. 2014; 26:2694–2701.
Article25. Jia L, Li H, Sun Y. Induction of p21-dependent senescence by a NAE inhibitor, MLN4924 as a mechanism of growth suppression. Neoplasia. 2011; 13:561–569.26. Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008; 451:1069–1075.
Article27. Tanida I. Autophagosome formation and molecular mechanism of autophagy. Antioxid Redox Signal. 2011; 14:2201–2214.
Article28. Kelekar A. Autophagy. Ann N Y Acad Sci. 2005; 1066:259–271.
Article29. Deegan S. The molecular characterization of ER stress-induced autophagy and cell death [PhD dissertation]. Ireland: NUI Galway;2012.30. Kim JH, Song HK. Swapping of interaction partners with ATG5 for autophagosome maturation. BMB Rep. 2015; 48:129–130.
Article31. Song L, Ma L, Chen G, Huang Y, Sun X, Jiang C, Liu H. Autophagy inhibitor 3-methyladenine enhances the sensitivity of nasopharyngeal carcinoma cells to chemotherapy and radiotherapy. J Cent South Univ (Med Sci). 2016; 41:9–18.32. Zhao Y, Xiong X, Jia L, Sun Y. Targeting Cullin-RING ligases by MLN4924 induces autophagy via modulating the HIF1-REDD1-TSC1-mTORC1-DEPTOR axis. Cell Death Dis. 2012; 3:e386.
Article33. Shen HM, Codogno P. Autophagic cell death: Loch Ness monster or endangered species? Autophagy. 2011; 7:457–465.
Article34. Codogno P, Meijer AJ. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 2005; 12:Suppl 2. 1509–1518.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Pan-Pim Kinase Inhibitor AZD1208 Suppresses Tumor Growth and Synergistically Interacts with Akt Inhibition in Gastric Cancer Cells
- Dual Roles of Autophagy and Their Potential Drugs for Improving Cancer Therapeutics
- Autophagy Inhibition Promotes Quercetin Induced Apoptosis in MG-63 Human Osteosarcoma cells
- Antiproliferative Activity of Gibbosic Acid H through Induction of G0/G1 Cell Cycle Arrest and Apoptosis in Human Lung Cancer Cells
- YM155, specific survivin inhibitor, can enhance artesunate-induced cytotoxicity in HCT116 colon cancer cells