Ann Lab Med.
2019 Mar;39(2):183-189. 10.3343/alm.2019.39.2.183.
An Excel Macro for Determining Allelic and Sequence Types of Bacterial Clones in Multilocus Sequence Typing
- Affiliations
-
- 1Department of Laboratory Medicine, Yonsei University College of Medicine, Seoul, Korea. kyunsky@yuhs.ac
- 2Research Institute of Bacterial Resistance, Yonsei University College of Medicine, Seoul, Korea.
- 3Division of Antimicrobial Resistance, National Institute of Health, Centers for Disease Control and Prevention, Cheongju, Korea.
- KMID: 2425974
- DOI: http://doi.org/10.3343/alm.2019.39.2.183
Abstract
- BACKGROUND
Multilocus sequence typing (MLST) was designed to overcome the low discriminatory power and poor reproducibility of previous molecular typing schemes, and it is useful for inter-laboratory, inter-regional, and inter-national comparison of pathogenic clones. MLST includes labor-intensive sequencing processes and meticulous allelic/sequence type (ST) determination processes, often prone to error. We developed a free automated MLST determination program (MLST typer) based on the Visual Basic for Applications macro, which runs on Microsoft Excel.
METHODS
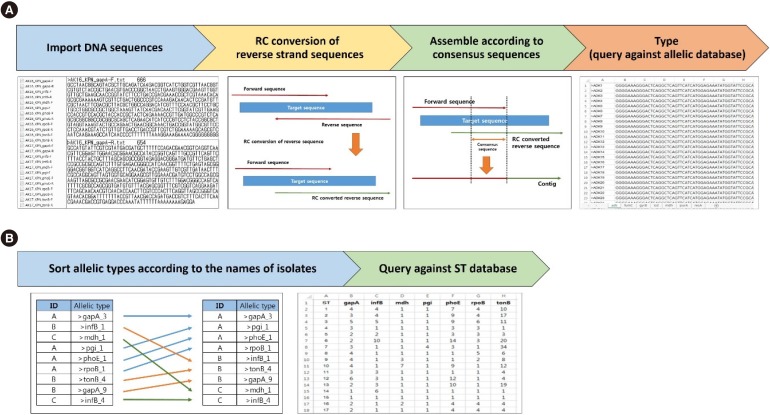
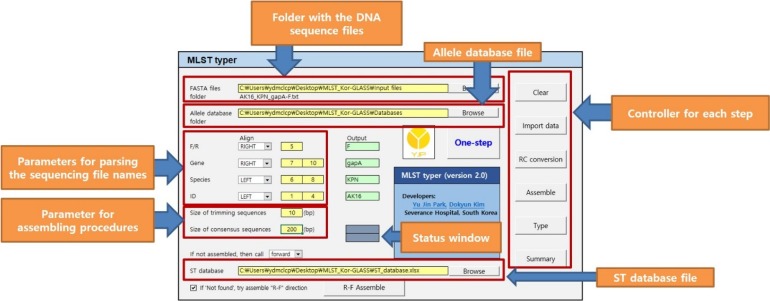
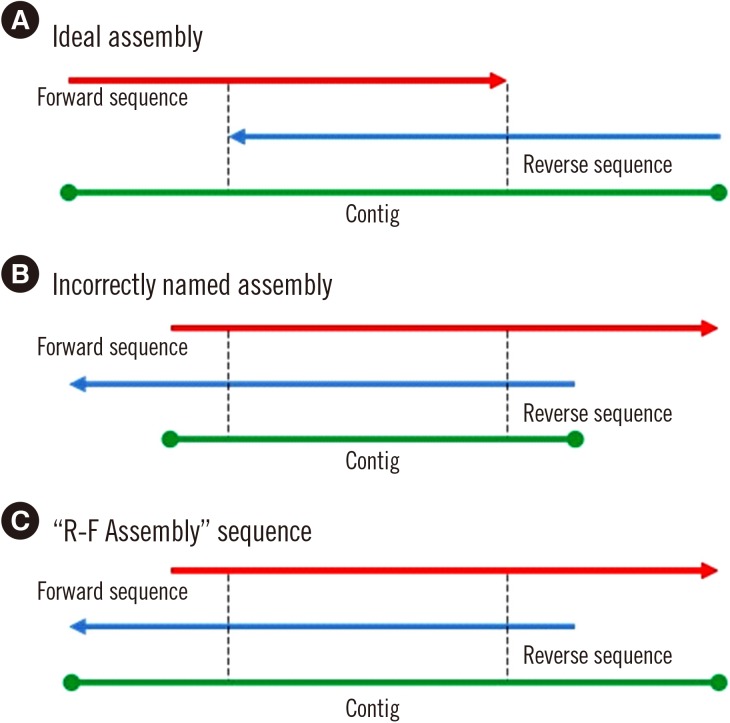
MLST typer imports sequence data in the FASTA format, converts reverse complement counterparts of the reverse sequences, assembles forward and reverse-complement converted sequences, and returns allelic numbers for each gene and ST of each isolate. To evaluate the performance of MLST typer, we tested the sequence data from 200 clinical isolates, each consisting of seven housekeeping gene sequences, with a total of 1,400 allelic number determinations. The results were compared with manual assessment.
RESULTS
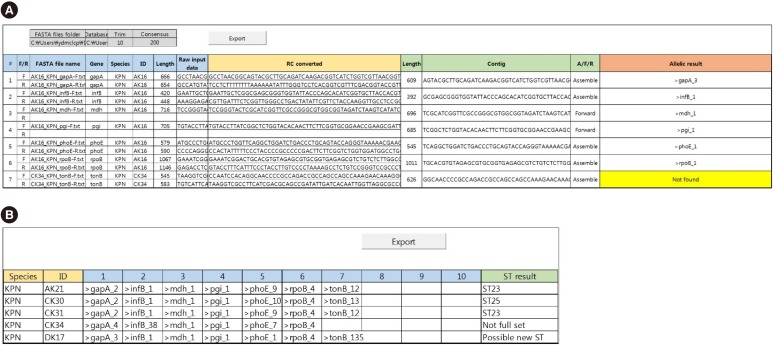
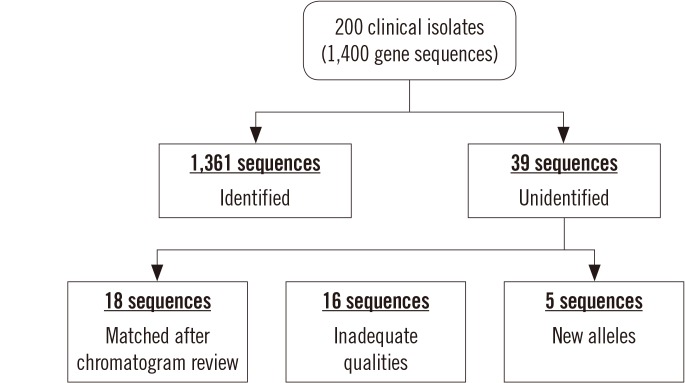
MLST typer comprises three worksheets: the Main page, Result page, and Summary page. The Main page console operates the process according to user-specified parameters. The Result and Summary pages provide the allelic type and ST determinations. It took approximately 12 minutes to analyze the sequence data from 200 clinical isolates. Compared with manual assessment, the rate of correct identification was 97.2% (1,361/1,400).
CONCLUSIONS
MLST typer can be widely used for epidemiological studies owing to its thoroughness in repetitive functions, good compatibility with FASTA type data files, and easy-to-understand outputs for allelic and ST determinations.
Keyword
MeSH Terms
Figure
Reference
-
1. MacCannell D. Bacterial strain typing. Clin Lab Med. 2013; 33:629–650. PMID: 23931842.2. Tenover FC, Arbeit RD, Goering RV. How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections: a review for healthcare epidemiologists. Molecular Typing Working Group of the Society for Healthcare Epidemiology of America. Infect Control Hosp Epidemiol. 1997; 18:426–439. PMID: 9181401.3. Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A. 1998; 95:3140–3145. PMID: 9501229.4. Maiden MC, Jansen van Rensburg MJ, Bray JE, Earle SG, Ford SA, Jolley KA, et al. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol. 2013; 11:728–736. PMID: 23979428.5. Sabat AJ, Budimir A, Nashev D, Sá-Leão R, van Dijl Jm, Laurent F, et al. Overview of molecular typing methods for outbreak detection and epidemiological surveillance. Euro Surveill. 2013; 18:20380. PMID: 23369389.6. Urwin R, Maiden MC. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 2003; 11:479–487. PMID: 14557031.7. Singh A, Goering RV, Simjee S, Foley SL, Zervos MJ. Application of molecular techniques to the study of hospital infection. Clin Microbiol Rev. 2006; 19:512–530. PMID: 16847083.8. Zhou H, Liu W, Qin T, Liu C, Ren H. Defining and evaluating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Klebsiella pneumoniae. Front Microbiol. 2017; 8:371. PMID: 28337187.9. Ilina EN, Oparina NY, Shitikov EA, Borovskaya AD, Govorun VM. Molecular surveillance of clinical Neisseria gonorrhoeae isolates in Russia. J Clin Microbiol. 2010; 48:3681–3689. PMID: 20660213.10. Bowden KE, Williams MM, Cassiday PK, Milton A, Pawloski L, Harrison M, et al. Molecular epidemiology of the pertussis epidemic in Washington State in 2012. J Clin Microbiol. 2014; 52:3549–3557. PMID: 25031439.11. Cantinelli T, Chenal-Francisque V, Diancourt L, Frezal L, Leclercq A, Wirth T, et al. ‘Epidemic clones’ of Listeria monocytogenes are widespread and ancient clonal groups. J Clin Microbiol. 2013; 51:3770–3779. PMID: 24006010.12. Aanensen DM, Spratt BG. The multilocus sequence typing network: mlst.net. Nucleic Acids Res. 2005; 33:W728–W733. PMID: 15980573.13. Jefferies J, Clarke SC, Diggle MA, Smith A, Dowson C, Mitchell T. Automated pneumococcal MLST using liquid-handling robotics and a capillary DNA sequencer. Mol Biotechnol. 2003; 24:303–308. PMID: 12777696.14. Clarke SC. Nucleotide sequence-based typing of bacteria and the impact of automation. Bioessays. 2002; 24:858–862. PMID: 12210523.15. Kardén-Lilja M, Vuopio J, Koskela M, Tissari P, Salmenlinna S. Molecular typing of vancomycin-resistant Enterococcus faecium with an automated repetitive sequence-based PCR microbial typing system compared with pulsed-field gel electrophoresis and multilocus sequence typing. Scand J Infect Dis. 2013; 45:350–356. PMID: 23163892.16. Sullivan CB, Jefferies JM, Diggle MA, Clarke SC. Automation of MLST using third-generation liquid-handling technology. Mol Biotechnol. 2006; 32:219–226. PMID: 16632888.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Characterization of a Vancomycin-resistant Enterococcus faecium Outbreak Caused by 2 Genetically Different Clones at a Neonatal Intensive Care Unit
- Modified Method of Multilocus Sequence Typing (MLST) for Serotyping in Salmonella Species
- Prevalence of Major Methicillin-Resistant Staphylococcus aureus Clones in Korea Between 2001 and 2008
- Expression of Sme Efflux Pumps and Multilocus Sequence Typing in Clinical Isolates of Stenotrophomonas maltophilia
- Multilocus Sequence Typing for Candida albicans Isolates from Candidemic Patients: Comparison with Southern Blot Hybridization and Pulsed-field Gel Electrophoresis Analysis