Cancer Res Treat.
2018 Oct;50(4):1378-1387. 10.4143/crt.2017.535.
Genetic Alterations among Korean Melanoma Patients Showing Tumor Heterogeneity: A Comparison between Primary Tumors and Corresponding Metastatic Lesions
- Affiliations
-
- 1Department of Dermatology, Cutaneous Biology Research Institute, Yonsei University College of Medicine, Seoul, Korea. karenroh@yuhs.ac
- 2Songdang Institute for Cancer Research, Yonsei Cancer Center, Yonsei University College of Medicine, Seoul, Korea. rha7655@yuhs.ac
- 3Division of Medical Oncology, Department of Internal Medicine, Yonsei University College of Medicine, Seoul, Korea.
- 4Brain Korea 21 Project for Medical Science, Seoul, Korea.
- KMID: 2424809
- DOI: http://doi.org/10.4143/crt.2017.535
Abstract
- PURPOSE
Melanoma is a highly heterogeneous neoplasm, composed of subpopulations of tumor cells with distinct molecular and biological phenotypes and genotypes. In this study, to determine the genetic heterogeneity between primary and metastatic melanoma in Korean melanoma patients, we evaluated several well-known genetic alterations of melanoma. In addition, to elucidate the clinical relevance of each genetic alteration and heterogeneity between primary and metastatic lesions, clinical features and patient outcome were collected.
MATERIALS AND METHODS
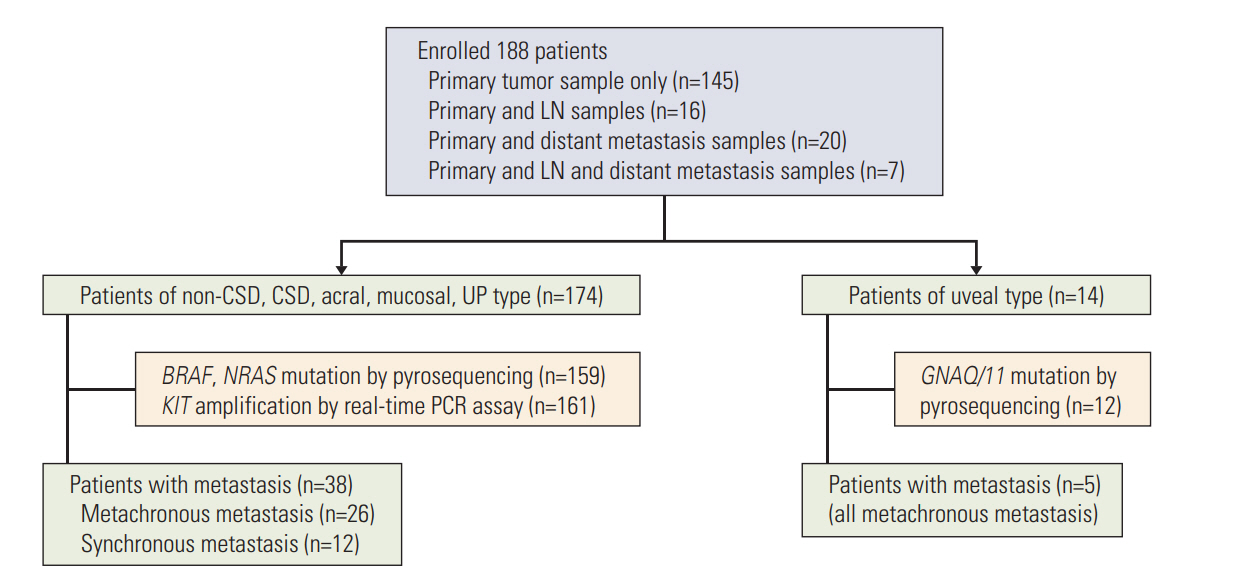
In addition to clinical data, BRAF, NRAS, GNAQ/11 mutation and KIT amplification data was acquired from an archived primary Korean melanoma cohort (KMC) of 188 patients. Among these patients, 43 patients were included for investigation of tumor heterogeneity between primary melanoma and its corresponding metastatic lesions.
RESULTS
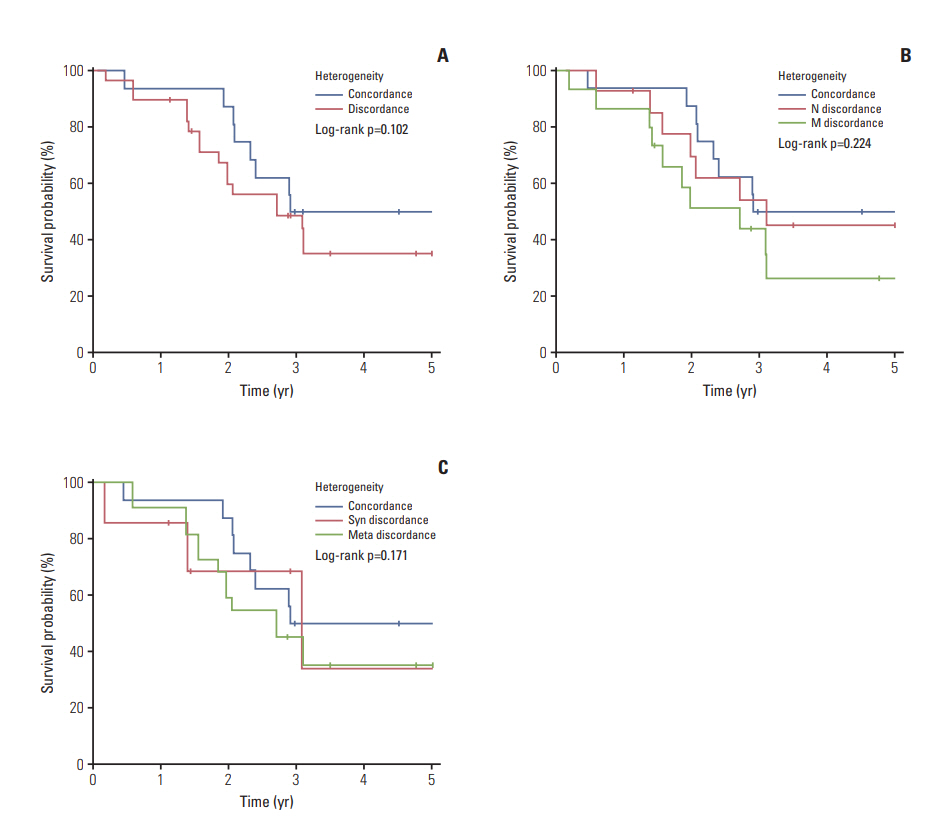
Overall incidence of genetic aberrations of the primary melanomas in KMC was 17.6% of BRAF V600, 12.6% of NRAS mutation, and 28.6% of KIT amplification. GNAQ/11 mutation was seen in 66.6% of the uveal melanoma patients. Patients with BRAF mutation were associated with advanced stage and correlated to poor prognosis (p < 0.01). Among 43 patients, 55.8% showed heterogeneity between primary and metastatic lesion. The frequency of BRAF mutation and KIT amplification significantly increased in the metastatic lesions compared to primary melanomas. GNAQ/11 mutation showed 100% homogeneity in uveal melanoma patients.
CONCLUSION
Our data demonstrated heterogeneity between primary melanomas and corresponding metastatic lesions for BRAF, NRAS mutation and KIT amplification. However, GNAQ/11 mutation was genetically homogeneous between primary and metastatic melanoma lesions in uveal melanoma.
Keyword
- Melanoma; Heterogeneity; BRAF; KIT; NRAS
MeSH Terms
Figure
Reference
-
References
1. Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013; 500:415–21.2. Fecher LA, Cummings SD, Keefe MJ, Alani RM. Toward a molecular classification of melanoma. J Clin Oncol. 2007; 25:1606–20.
Article3. Miller AJ, Mihm MC Jr. Melanoma. N Engl J Med. 2006; 355:51–65.
Article4. Cancer Genome Atlas Network. Genomic classification of cutaneous melanoma. Cell. 2015; 161:1681–96.5. Kong Y, Si L, Zhu Y, Xu X, Corless CL, Flaherty KT, et al. Large-scale analysis of KIT aberrations in Chinese patients with melanoma. Clin Cancer Res. 2011; 17:1684–91.
Article6. Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, et al. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010; 141:583–94.
Article7. Yancovitz M, Litterman A, Yoon J, Ng E, Shapiro RL, Berman RS, et al. Intra- and inter-tumor heterogeneity of BRAF (V600E) mutations in primary and metastatic melanoma. PLoS One. 2012; 7:e29336.8. Yun J, Lee J, Jang J, Lee EJ, Jang KT, Kim JH, et al. KIT amplification and gene mutations in acral/mucosal melanoma in Korea. APMIS. 2011; 119:330–5.
Article9. Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010; 363:2191–9.10. Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002; 417:949–54.
Article11. Flaherty KT, Hodi FS, Bastian BC. Mutation-driven drug development in melanoma. Curr Opin Oncol. 2010; 22:178–83.
Article12. Hiley C, de Bruin EC, McGranahan N, Swanton C. Deciphering intratumor heterogeneity and temporal acquisition of driver events to refine precision medicine. Genome Biol. 2014; 15:453.
Article13. Woodman SE, Lazar AJ, Aldape KD, Davies MA. New strategies in melanoma: molecular testing in advanced disease. Clin Cancer Res. 2012; 18:1195–200.
Article14. Greaves WO, Verma S, Patel KP, Davies MA, Barkoh BA, Galbincea JM, et al. Frequency and spectrum of BRAF mutations in a retrospective, single-institution study of 1112 cases of melanoma. J Mol Diagn. 2013; 15:220–6.
Article15. Oh CM, Cho H, Won YJ, Kong HJ, Roh YH, Jeong KH, et al. Nationwide trends in the incidence of melanoma and nonmelanoma skin cancers from 1999 to 2014 in South Korea. Cancer Res Treat. 2018; 50:729–37.
Article16. Thompson JF, Scolyer RA, Kefford RF. Cutaneous melanoma. Lancet. 2005; 365:687–701.
Article17. Balch CM, Soong SJ, Gershenwald JE, Thompson JF, Reintgen DS, Cascinelli N, et al. Prognostic factors analysis of 17,600 melanoma patients: validation of the American Joint Committee on Cancer melanoma staging system. J Clin Oncol. 2001; 19:3622–34.
Article18. Long GV, Menzies AM, Nagrial AM, Haydu LE, Hamilton AL, Mann GJ, et al. Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. J Clin Oncol. 2011; 29:1239–46.
Article19. Fecher LA, Amaravadi RK, Flaherty KT. The MAPK pathway in melanoma. Curr Opin Oncol. 2008; 20:183–9.
Article20. Edlundh-Rose E, Egyhazi S, Omholt K, Mansson-Brahme E, Platz A, Hansson J, et al. NRAS and BRAF mutations in melanoma tumours in relation to clinical characteristics: a study based on mutation screening by pyrosequencing. Melanoma Res. 2006; 16:471–8.
Article21. Ugurel S, Thirumaran RK, Bloethner S, Gast A, Sucker A, Mueller-Berghaus J, et al. B-RAF and N-RAS mutations are preserved during short time in vitro propagation and differentially impact prognosis. PLoS One. 2007; 2:e236.
Article22. Bauer J, Buttner P, Murali R, Okamoto I, Kolaitis NA, Landi MT, et al. BRAF mutations in cutaneous melanoma are independently associated with age, anatomic site of the primary tumor, and the degree of solar elastosis at the primary tumor site. Pigment Cell Melanoma Res. 2011; 24:345–51.
Article23. Ellerhorst JA, Greene VR, Ekmekcioglu S, Warneke CL, Johnson MM, Cooke CP, et al. Clinical correlates of NRAS and BRAF mutations in primary human melanoma. Clin Cancer Res. 2011; 17:229–35.24. Almendro V, Marusyk A, Polyak K. Cellular heterogeneity and molecular evolution in cancer. Annu Rev Pathol. 2013; 8:277–302.
Article25. Thomas NE, Alexander A, Edmiston SN, Parrish E, Millikan RC, Berwick M, et al. Tandem BRAF mutations in primary invasive melanomas. J Invest Dermatol. 2004; 122:1245–50.
Article26. Patton EE, Widlund HR, Kutok JL, Kopani KR, Amatruda JF, Murphey RD, et al. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr Biol. 2005; 15:249–54.
Article27. Colombino M, Capone M, Lissia A, Cossu A, Rubino C, De Giorgi V, et al. BRAF/NRAS mutation frequencies among primary tumors and metastases in patients with melanoma. J Clin Oncol. 2012; 30:2522–9.
Article28. Greene VR, Johnson MM, Grimm EA, Ellerhorst JA. Frequencies of NRAS and BRAF mutations increase from the radial to the vertical growth phase in cutaneous melanoma. J Invest Dermatol. 2009; 129:1483–8.
Article29. Lin J, Goto Y, Murata H, Sakaizawa K, Uchiyama A, Saida T, et al. Polyclonality of BRAF mutations in primary melanoma and the selection of mutant alleles during progression. Br J Cancer. 2011; 104:464–8.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Metastatic Malignant Melanoma Representing a Multiple Mesenteric Cystic Tumor: A Case Report
- p53 Expression in Malignant Melanoma and Melanocytic Nevus
- The Significance of Bone Scan in Malignant Bone Tumors
- Diagnosis and management of primary malignant melanoma of the lung: a case report
- Unknow Primary Melanoma