Yonsei Med J.
2018 Mar;59(2):211-218. 10.3349/ymj.2018.59.2.211.
Peptide Nucleic Acid Clamping and Direct Sequencing in the Detection of Oncogenic Alterations in Lung Cancer: Systematic Review and Meta-Analysis
- Affiliations
-
- 1Division of Pulmonary and Critical Care Medicine, Department of Internal Medicine, Kangbuk Samsung Hospital, Sungkyunkwan University School of Medicine, Seoul, Korea.
- 2Department of Internal Medicine, Jeju National University Hospital, Jeju National University School of Medicine, Jeju, Korea. lovlet@paran.com
- KMID: 2418783
- DOI: http://doi.org/10.3349/ymj.2018.59.2.211
Abstract
- PURPOSE
Molecular testing in non-small cell lung cancer (NSCLC) aids in identifying oncogenic alterations. The aim of this study was to compare the rates of detection of oncogenic alterations and responsiveness to epidermal growth factor receptor-tyrosine kinase inhibitors (EGFR-TKIs) according to EGFR mutation status as determined by peptide nucleic acid (PNA) clamping or direct sequencing (DS).
MATERIALS AND METHODS
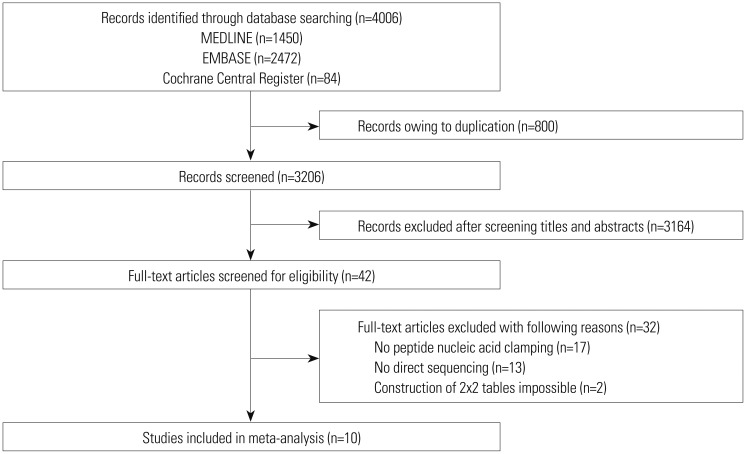
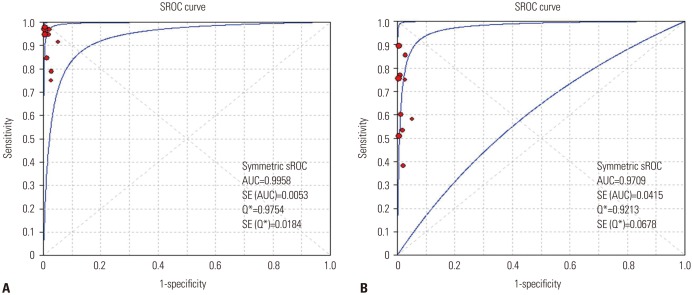
We performed a systematic literature search using MEDLINE, EMBASE, and the Cochrane Central Register. Data from included studies were pooled to yield summary sensitivity, specificity, positive and negative likelihood ratios, diagnostic odds ratio, and receiver operating characteristic curves. A meta-regression analysis was conducted to identify potential sources of heterogeneity between selected studies.
RESULTS
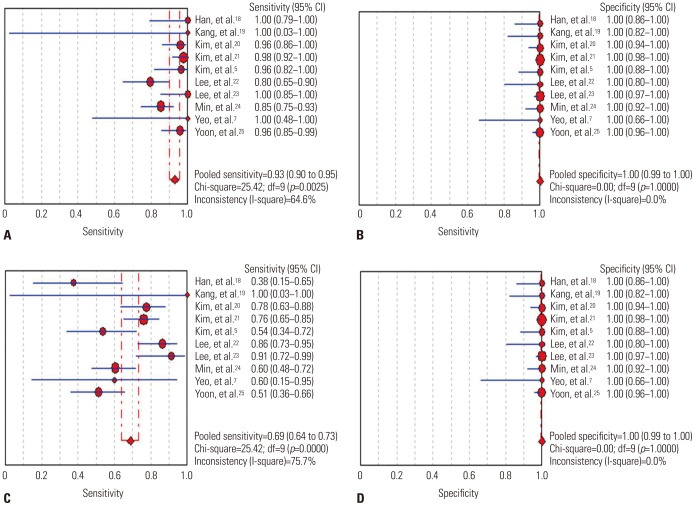
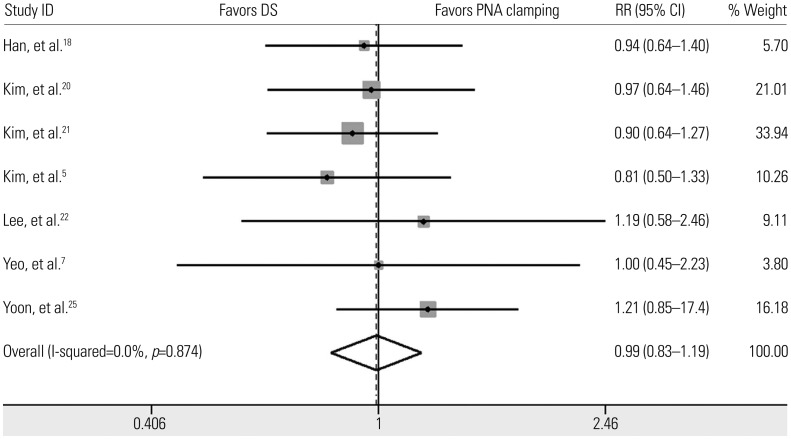
We identified 10 studies comprising 924 patients. Oncogenic alterations were detected in 340 of 924 cases (36.8%) with PNA clamping and in 250 of 924 (27.1%) with DS. The pooled sensitivities of PNA clamping and DS were 0.93 [95% confidence interval (CI): 0.90−0.95] and 0.69 (95% CI: 0.64−0.73), respectively. According to meta-regression analysis, none of the covariates were found to be significant sources of heterogeneity. With respect to treatment responses to EGFR-TKIs, there was no significant difference therein between EGFR mutations detected by PNA clamping and DS (53.4% vs. 50.8%; risk ratio, 0.99; 95% CI 0.83−1.19; p=0.874).
CONCLUSION
We demonstrated that PNA clamping has a higher sensitivity than DS for detecting oncogenic alterations in NSCLC. Our findings suggest that PNA clamping is a more useful method for clinical practice.
MeSH Terms
-
Antineoplastic Agents/therapeutic use
Carcinoma, Non-Small-Cell Lung/drug therapy/*genetics
Constriction
Humans
Lung Neoplasms/*genetics
Molecular Diagnostic Techniques
Mutation
Peptide Nucleic Acids/*genetics
Protein Kinase Inhibitors/*therapeutic use
Receptor Protein-Tyrosine Kinases/*genetics
Receptor, Epidermal Growth Factor/*genetics
Sensitivity and Specificity
Sequence Analysis
Sequence Analysis, DNA
Translocation, Genetic
Antineoplastic Agents
Peptide Nucleic Acids
Protein Kinase Inhibitors
Receptor Protein-Tyrosine Kinases
Receptor, Epidermal Growth Factor
Figure
Reference
-
1. Reck M, Rabe KF. Precision diagnosis and treatment for advanced non-small-cell lung cancer. N Engl J Med. 2017; 377:849–861. PMID: 28854088.
Article2. Won JK, Keam B, Koh J, Cho HJ, Jeon YK, Kim TM, et al. Concomitant ALK translocation and EGFR mutation in lung cancer: a comparison of direct sequencing and sensitive assays and the impact on responsiveness to tyrosine kinase inhibitor. Ann Oncol. 2015; 26:348–354. PMID: 25403583.
Article3. Lindeman NI, Cagle PT, Beasley MB, Chitale DA, Dacic S, Giaccone G, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. Arch Pathol Lab Med. 2013; 137:828–860. PMID: 23551194.
Article4. Pao W, Ladanyi M. Epidermal growth factor receptor mutation testing in lung cancer: searching for the ideal method. Clin Cancer Res. 2007; 13:4954–4955. PMID: 17785543.5. Kim HS, Sung JS, Yang SJ, Kwon NJ, Jin L, Kim ST, et al. Predictive efficacy of low burden EGFR mutation detected by next-generation sequencing on response to EGFR tyrosine kinase inhibitors in non-small-cell lung carcinoma. PLoS One. 2013; 8:e81975. PMID: 24376508.
Article6. Lee KY, Kim HJ, Kim SJ, Yoo GH, Kim WD, Oh SY, et al. PNA-mediated PCR clamping for the detection of EGFR mutations in non-small cell lung cancer. Tuberc Respir Dis. 2010; 69:271–278.
Article7. Yeo CD, Kim JW, Kim KH, Ha JH, Rhee CK, Kim SJ, et al. Detection and comparison of EGFR mutations in matched tumor tissues, cell blocks, pleural effusions, and sera from patients with NSCLC with malignant pleural effusion, by PNA clamping and direct sequencing. Lung Cancer. 2013; 81:207–212. PMID: 23726527.
Article8. Sakurada A, Lara-Guerra H, Liu N, Shepherd FA, Tsao MS. Tissue heterogeneity of EGFR mutation in lung adenocarcinoma. J Thorac Oncol. 2008; 3:527–529. PMID: 18449007.
Article9. Taniguchi K, Okami J, Kodama K, Higashiyama M, Kato K. Intratumor heterogeneity of epidermal growth factor receptor mutations in lung cancer and its correlation to the response to gefitinib. Cancer Sci. 2008; 99:929–935. PMID: 18325048.
Article10. Yeh YC, Chou TY. Pulmonary adenocarcinoma with microcystic histology and intratumoral heterogeneity of EGFR gene polymorphism. Histopathology. 2010; 57:112–120. PMID: 20653783.
Article11. Yatabe Y, Matsuo K, Mitsudomi T. Heterogeneous distribution of EGFR mutations is extremely rare in lung adenocarcinoma. J Clin Oncol. 2011; 29:2972–2977. PMID: 21730270.
Article12. Moher D, Liberati A, Tetzlaff J, Altman DG. PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009; 151:264–269. PMID: 19622511.
Article13. Whiting P, Rutjes AW, Reitsma JB, Bossuyt PM, Kleijnen J. The development of QUADAS: a tool for the quality assessment of studies of diagnostic accuracy included in systematic reviews. BMC Med Res Methodol. 2003; 3:25. PMID: 14606960.
Article14. Reitsma JB, Glas AS, Rutjes AW, Scholten RJ, Bossuyt PM, Zwinderman AH. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J Clin Epidemiol. 2005; 58:982–990. PMID: 16168343.
Article15. Thompson SG. Why sources of heterogeneity in meta-analysis should be investigated. BMJ. 1994; 309:1351–1355. PMID: 7866085.16. Lijmer JG, Mol BW, Heisterkamp S, Bonsel GJ, Prins MH, van der Meulen JH, et al. Empirical evidence of design-related bias in studies of diagnostic tests. JAMA. 1999; 282:1061–1066. PMID: 10493205.
Article17. Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol. 2005; 58:882–893. PMID: 16085191.
Article18. Han HS, Lim SN, An JY, Lee KM, Choe KH, Lee KH, et al. Detection of EGFR mutation status in lung adenocarcinoma specimens with different proportions of tumor cells using two methods of differential sensitivity. J Thorac Oncol. 2012; 7:355–364. PMID: 22157369.
Article19. Kang JY, Park CK, Yeo CD, Lee HY, Rhee CK, Kim SJ, et al. Comparison of PNA clamping and direct sequencing for detecting KRAS mutations in matched tumour tissue, cell block, pleural effusion and serum from patients with malignant pleural effusion. Respirology. 2015; 20:138–146. PMID: 25302858.20. Kim HJ, Kim WS, Shin KC, Lee GH, Kim MJ, Lee JE, et al. Comparative analysis of peptide nucleic acid (PNA)-mediated real-time PCR clamping and DNA direct sequencing for EGFR mutation detection. Tuberc Respir Dis. 2011; 70:21–27.
Article21. Kim HJ, Lee KY, Kim YC, Kim KS, Lee SY, Jang TW, et al. Detection and comparison of peptide nucleic acid-mediated real-time polymerase chain reaction clamping and direct gene sequencing for epidermal growth factor receptor mutations in patients with non-small cell lung cancer. Lung Cancer. 2012; 75:321–325. PMID: 21930325.
Article22. Lee HJ, Xu X, Kim H, Jin Y, Sun P, Kim JE, et al. Comparison of direct sequencing, PNA clamping-real time polymerase chain reaction, and pyrosequencing methods for the detection of EGFR mutations in non-small cell lung carcinoma and the correlation with clinical responses to EGFR tyrosine kinase inhibitor treatment. Korean J Pathol. 2013; 47:52–60. PMID: 23483646.23. Lee B, Lee B, Han G, Kwon MJ, Han J, Choi YL. KRAS mutation detection in non-small cell lung cancer using a peptide nucleic acid-mediated polymerase chain reaction clamping method and comparative validation with next-generation sequencing. Korean J Pathol. 2014; 48:100–107. PMID: 24868222.24. Min KW, Kim WS, Jang SJ, Choi YD, Chang S, Jung SH, et al. Comparison of EGFR mutation detection between the tissue and cytology using direct sequencing, pyrosequencing and peptide nucleic acid clamping in lung adenocarcinoma: Korean multicentre study. QJM. 2016; 109:167–173. PMID: 26031706.
Article25. Yoon SH, Choi YD, Oh IJ, Kim KS, Choi H, Chang J, et al. Peptide nucleic acid clamping versus direct sequencing for the detection of EGFR gene mutation in patients with non-small cell lung cancer. Cancer Res Treat. 2015; 47:661–669. PMID: 25715768.26. Endo K, Konishi A, Sasaki H, Takada M, Tanaka H, Okumura M, et al. Epidermal growth factor receptor gene mutation in non-small cell lung cancer using highly sensitive and fast TaqMan PCR assay. Lung Cancer. 2005; 50:375–384. PMID: 16199108.
Article27. Pirker R, Herth FJ, Kerr KM, Filipits M, Taron M, Gandara D, et al. Consensus for EGFR mutation testing in non-small cell lung cancer: results from a European workshop. J Thorac Oncol. 2010; 5:1706–1713. PMID: 20871269.
Article28. Dinnes J, Deeks J, Kirby J, Roderick P. A methodological review of how heterogeneity has been examined in systematic reviews of diagnostic test accuracy. Health Technol Assess. 2005; 9(12):1–113.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- KRAS Mutation Detection in Non-small Cell Lung Cancer Using a Peptide Nucleic Acid-Mediated Polymerase Chain Reaction Clamping Method and Comparative Validation with Next-Generation Sequencing
- Peptide Nucleic Acid Clamping Versus Direct Sequencing for the Detection of EGFR Gene Mutation in Patients with Non-small Cell Lung Cancer
- Comparative Analysis of Peptide Nucleic Acid (PNA)-Mediated Real-Time PCR Clamping and DNA Direct Sequencing for EGFR Mutation Detection
- Comparison of Direct Sequencing, PNA Clamping-Real Time Polymerase Chain Reaction, and Pyrosequencing Methods for the Detection of EGFR Mutations in Non-small Cell Lung Carcinoma and the Correlation with Clinical Responses to EGFR Tyrosine Kinase Inhibitor Treatment
- Rapid and Sensitive Detection of KRAS Mutation by Peptide Nucleic Acid-based Real-time PCR Clamping: A Comparison with Direct Sequencing between Fresh Tissue and Formalin-fixed and Paraffin Embedded Tissue of Colorectal Cancer