Cancer Res Treat.
2018 Jul;50(3):936-949. 10.4143/crt.2017.302.
MicroRNA-373 Inhibits Cell Proliferation and Invasion via Targeting BRF2 in Human Non-small Cell Lung Cancer A549 Cell Line
- Affiliations
-
- 1Department of Thoracic Surgery, The First Affiliated Hospital of Harbin Medical University, Harbin, China. wangju6158@sina.com
- 2The Central Operating Room, The First Affiliated Hospital of Harbin Medical University, Harbin, China.
- KMID: 2417882
- DOI: http://doi.org/10.4143/crt.2017.302
Abstract
- PURPOSE
The purpose of this study was to investigate the biological role and mechanism of miR-373 targeting of TFIIB-related factor 2 (BRF2) in the regulation of non-small cell lung cancer (NSCLC) cells.
MATERIALS AND METHODS
miRNA microarray chip analysis of four paired NSCLC and adjacent non-tumor tissues was performed. Quantitative real-time polymerase chain reaction (qRT-PCR) andwestern blotting were used to detect the expression levels of miR-373 and BRF2 in NSCLC tissues and cell lines. The dual-luciferase reporter method was performed to determine if BRF2 is a target of miR-373. MTT, wound-healing, Transwell, and flow cytometric assays were conducted to examine the proliferation, migration, invasion, and cell cycle progression of NSCLC A549 cells, respectively; western blotting was used to detect the expression of epithelial-mesenchymal transition (EMT)-related proteins.
RESULTS
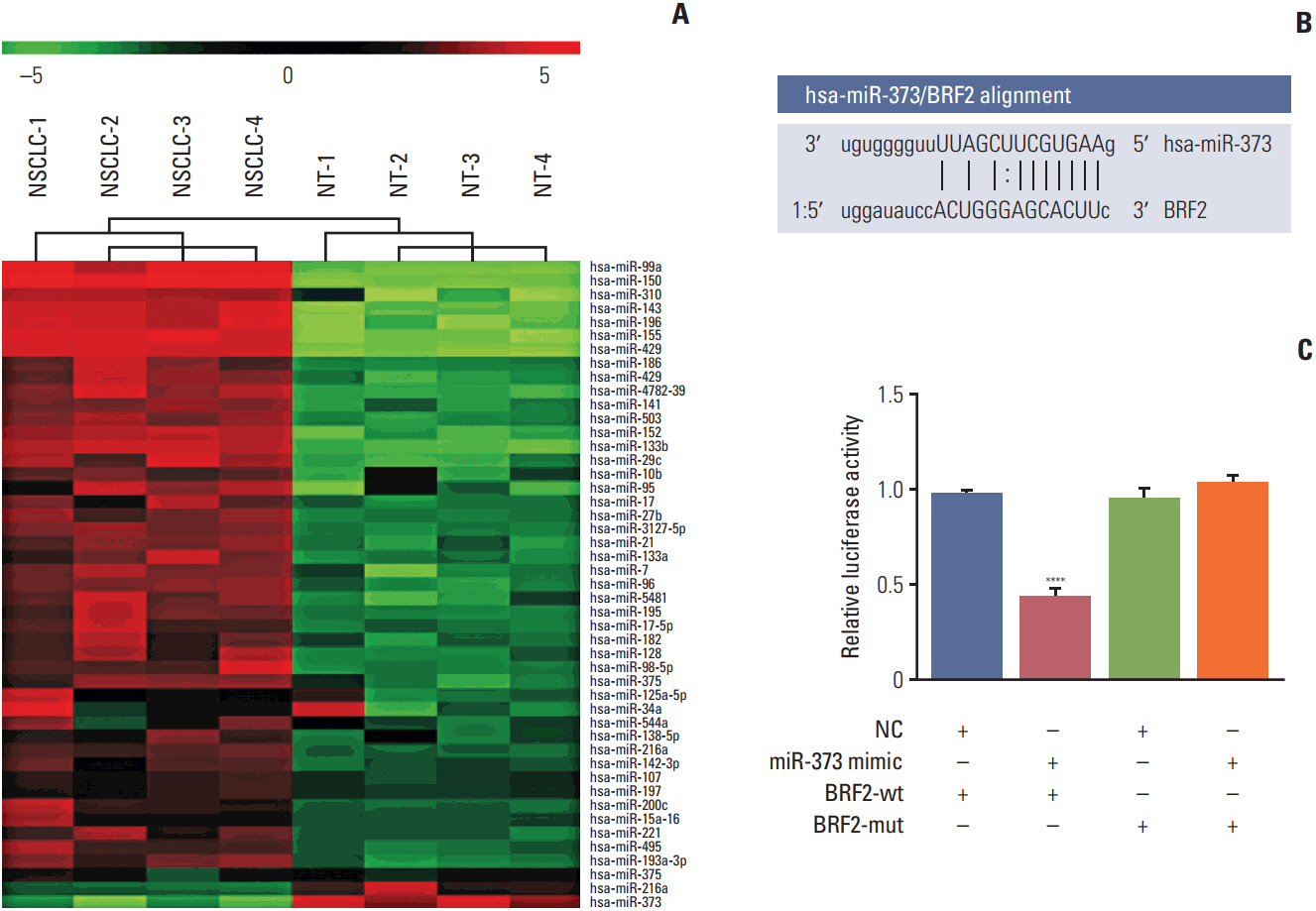
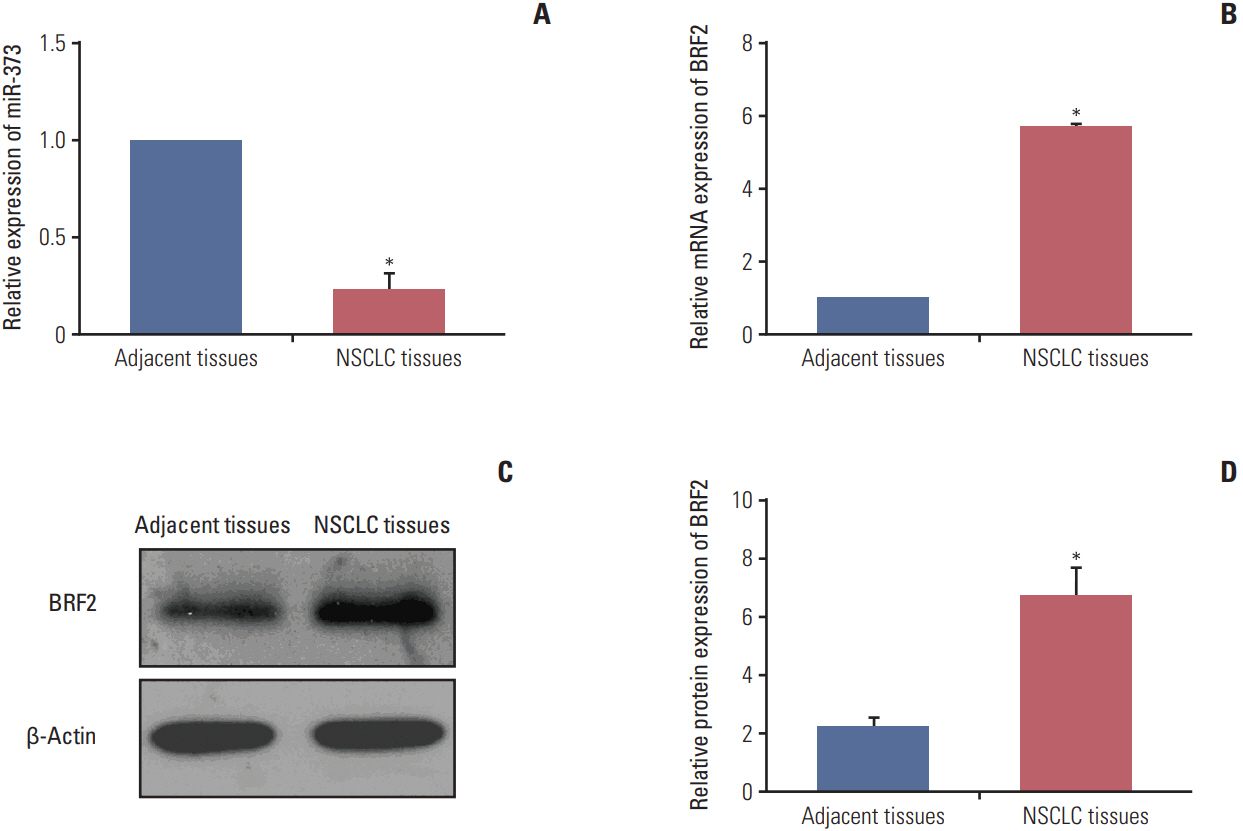
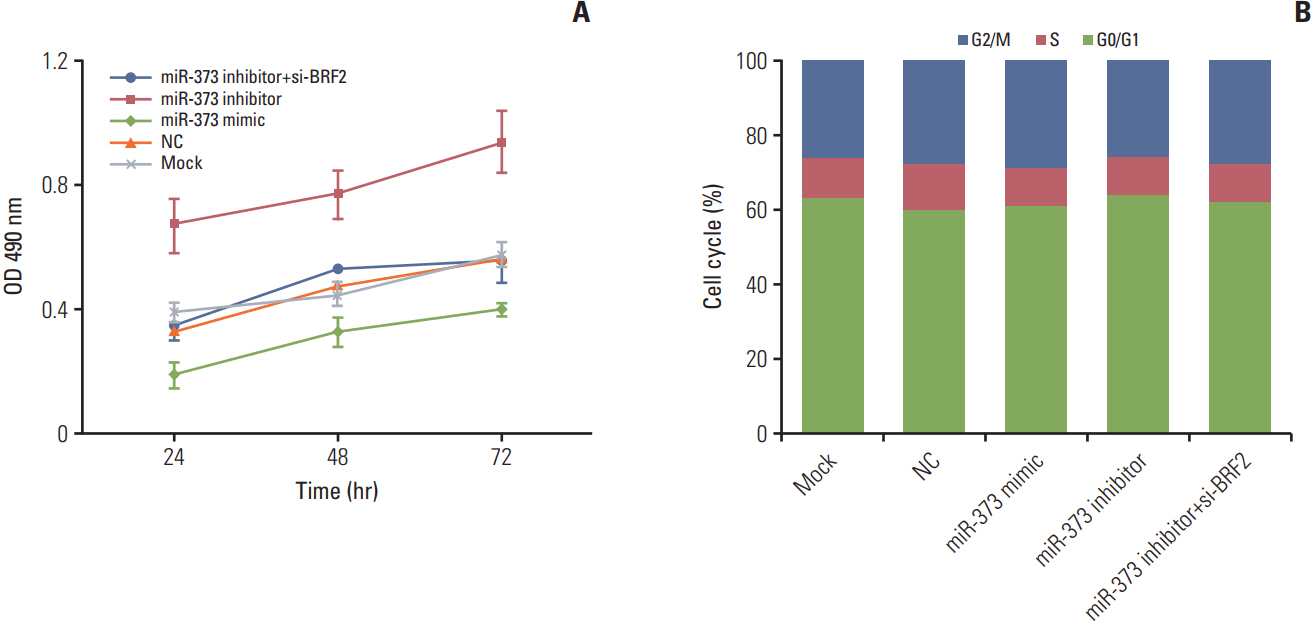
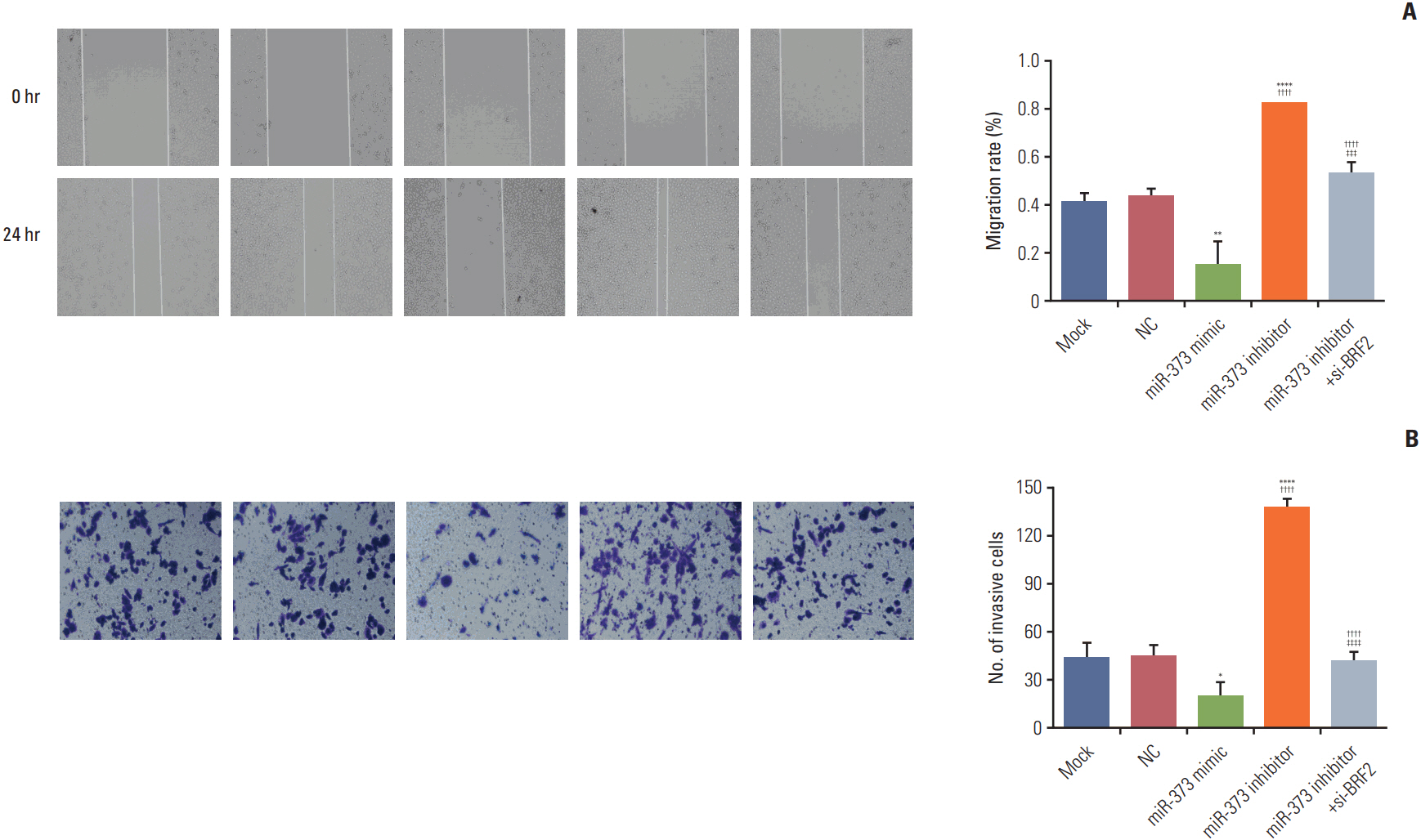
The miRNA microarray chip analysis demonstrated that miR-373 was down-regulated in NSCLC tissues, and this result was confirmed by qRT-PCR. Additionally, miR-373 was confirmed to target BRF2. Moreover, miR-373 expression was inversely correlated with BRF2 expression in NSCLC tissues and cell lines; both miR-373 down-regulation and BRF2 up-regulation were strongly associated with the clinicopathological features and prognosis of NSCLC patients. In vitro, overexpression of miR-373 markedly inhibited cell proliferation, migration, and invasion; up-regulated the expression of E-cadherin; and down-regulated the expression of N-cadherin and Snail in A549 cell. Knockdown BRF2 by siRNA resulted in effects similar to those caused by overexpression of miR-373.
CONCLUSION
MiR-373 is decreased in NSCLC, and overexpression of miR-373 can suppress cell EMT, and inhibit the proliferation, migration, and invasion of NSCLC A549 cells by targeting BRF2.
Keyword
MeSH Terms
-
Blotting, Western
Cadherins
Carcinoma, Non-Small-Cell Lung*
Cell Cycle
Cell Line*
Cell Migration Inhibition
Cell Proliferation*
Down-Regulation
Epithelial-Mesenchymal Transition
Humans*
In Vitro Techniques
Methods
MicroRNAs
Prognosis
Real-Time Polymerase Chain Reaction
RNA, Small Interfering
Snails
Up-Regulation
Cadherins
MicroRNAs
RNA, Small Interfering
Figure
Reference
-
References
1. Wang F, Meng F, Wang L, Wong SC, Cho WC, Chan LW. Associations of mRNA:microRNA for the shared downstream molecules of EGFR and alternative tyrosine kinase receptors in non-small cell lung cancer. Front Genet. 2016; 7:173.
Article2. Mahdavifar N, Ghoncheh M, Pakzad R, Momenimovahed Z, Salehiniya H. Epidemiology, incidence and mortality of bladder cancer and their relationship with the development index in the world. Asian Pac J Cancer Prev. 2016; 17:381–6.
Article3. Souza MC, Cruz OG, Vasconcelos AG. Factors associated with disease-specific survival of patients with non-small cell lung cancer. J Bras Pneumol. 2016; 42:317–25.
Article4. Bao W, Greenwold MJ, Sawyer RH. Expressed miRNAs target feather related mRNAs involved in cell signaling, cell adhesion and structure during chicken epidermal development. Gene. 2016; 591:393–402.
Article5. Peterson SM, Thompson JA, Ufkin ML, Sathyanarayana P, Liaw L, Congdon CB. Common features of microRNA target prediction tools. Front Genet. 2014; 5:23.
Article6. Shenoy A, Blelloch RH. Regulation of microRNA function in somatic stem cell proliferation and differentiation. Nat Rev Mol Cell Biol. 2014; 15:565–76.
Article7. Li S, Ma Y, Hou X, Liu Y, Li K, Xu S, et al. MiR-185 acts as a tumor suppressor by targeting AKT1 in non-small cell lung cancer cells. Int J Clin Exp Pathol. 2015; 8:11854–62.8. Ma N, Zhang W, Qiao C, Luo H, Zhang X, Liu D, et al. The tumor suppressive role of miRNA-509-5p by targeting FOXM1 in non-small cell lung cancer. Cell Physiol Biochem. 2016; 38:1435–46.
Article9. Seol HS, Akiyama Y, Shimada S, Lee HJ, Kim TI, Chun SM, et al. Epigenetic silencing of microRNA-373 to epithelial-mesenchymal transition in non-small cell lung cancer through IRAK2 and LAMP1 axes. Cancer Lett. 2014; 353:232–41.
Article10. Yan GR, Xu SH, Tan ZL, Liu L, He QY. Global identification of miR-373-regulated genes in breast cancer by quantitative proteomics. Proteomics. 2011; 11:912–20.
Article11. Koo CX, Kobiyama K, Shen YJ, LeBert N, Ahmad S, Khatoo M, et al. RNA polymerase III regulates cytosolic RNA:DNA hybrids and intracellular microRNA expression. J Biol Chem. 2015; 290:7463–73.
Article12. Gouge J, Satia K, Guthertz N, Widya M, Thompson AJ, Cousin P, et al. Redox signaling by the RNA polymerase III TFIIBrelated factor Brf2. Cell. 2015; 163:1375–87.
Article13. Saxena A, Ma B, Schramm L, Hernandez N. Structure-function analysis of the human TFIIB-related factor II protein reveals an essential role for the C-terminal domain in RNA polymerase III transcription. Mol Cell Biol. 2005; 25:9406–18.
Article14. Lu M, Tian H, Yue W, Li L, Li S, Qi L, et al. Overexpression of TFIIB-related factor 2 is significantly correlated with tumor angiogenesis and poor survival in patients with esophageal squamous cell cancer. Med Oncol. 2013; 30:553.
Article15. Cabarcas S, Schramm L. RNA polymerase III transcription in cancer: the BRF2 connection. Mol Cancer. 2011; 10:47.
Article16. Lu M, Tian H, Yue W, Li L, Li S, Qi L, et al. TFIIB-related factor 2 over expression is a prognosis marker for early-stage nonsmall cell lung cancer correlated with tumor angiogenesis. PLoS One. 2014; 9:e88032.
Article17. Lockwood WW, Chari R, Coe BP, Thu KL, Garnis C, Malloff CA, et al. Integrative genomic analyses identify BRF2 as a novel lineage-specific oncogene in lung squamous cell carcinoma. PLoS Med. 2010; 7:e1000315.
Article18. Wrona A, Jassem J. The new TNM classification in lung cancer. Pneumonol Alergol Pol. 2010; 78:407–17.19. Gallagher MF, Heffron CC, Laios A, O'Toole SA, Ffrench B, Smyth PC, et al. Suppression of cancer stemness p21-regulating mRNA and microRNA signatures in recurrent ovarian cancer patient samples. J Ovarian Res. 2012; 5:2.
Article20. Wang S, Ma G, Zhu H, Lv C, Chu H, Tong N, et al. miR-107 regulates tumor progression by targeting NF1 in gastric cancer. Sci Rep. 2016; 6:36531.
Article21. Eyking A, Reis H, Frank M, Gerken G, Schmid KW, Cario E. MiR-205 and miR-373 are associated with aggressive human mucinous colorectal cancer. PLoS One. 2016; 11:e0156871.
Article22. Zhang Y, Zhao FJ, Chen LL, Wang LQ, Nephew KP, Wu YL, et al. MiR-373 targeting of the Rab22a oncogene suppresses tumor invasion and metastasis in ovarian cancer. Oncotarget. 2014; 5:12291–303.
Article23. Tanaka T, Arai M, Wu S, Kanda T, Miyauchi H, Imazeki F, et al. Epigenetic silencing of microRNA-373 plays an important role in regulating cell proliferation in colon cancer. Oncol Rep. 2011; 26:1329–35.
Article24. Chen Y, Luo J, Tian R, Sun H, Zou S. miR-373 negatively regulates methyl-CpG-binding domain protein 2 (MBD2) in hilar cholangiocarcinoma. Dig Dis Sci. 2011; 56:1693–701.
Article25. Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Adv Exp Med Biol. 2007; 604:17–46.
Article26. Wang Z, Wu YL, Zhang GC, Zhou Q, Xu CR, Guo AL. EGFR/KRAS mutations and gefitinib therapy in Chinese NSCLC patients. Onkologie. 2008; 31:174–8.
Article27. Jing SY, Jing SQ, Liu LL, Xu LF, Zhang F, Gao JL. Downexpression of miR-373 predicts poor prognosis of glioma and could be a potential therapeutic target. Eur Rev Med Pharmacol Sci. 2017; 21:2421–5.28. Eichelser C, Flesch-Janys D, Chang-Claude J, Pantel K, Schwarzenbach H. Deregulated serum concentrations of circulating cell-free microRNAs miR-17, miR-34a, miR-155, and miR-373 in human breast cancer development and progression. Clin Chem. 2013; 59:1489–96.
Article29. Koo J, Cabarcas-Petroski S, Petrie JL, Diette N, White RJ, Schramm L. Induction of proto-oncogene BRF2 in breast cancer cells by the dietary soybean isoflavone daidzein. BMC Cancer. 2015; 15:905.
Article30. Papiewska-Pajak I, Kowalska MA, Boncela J. Expression and activity of SNAIL transcription factor during epithelial to mesenchymal transition (EMT) in cancer progression. Postepy Hig Med Dosw (Online). 2016; 70:968–80.31. Gravdal K, Halvorsen OJ, Haukaas SA, Akslen LA. A switch from E-cadherin to N-cadherin expression indicates epithelial to mesenchymal transition and is of strong and independent importance for the progress of prostate cancer. Clin Cancer Res. 2007; 13:7003–11.
Article32. Wang Y, Shi J, Chai K, Ying X, Zhou BP. The role of Snail in EMT and tumorigenesis. Curr Cancer Drug Targets. 2013; 13:963–72.
Article33. Tian Y, Lu M, Yue W, Li L, Li S, Gao C, et al. TFIIB-related factor 2 is associated with poor prognosis of nonsmall cell lung cancer patients through promoting tumor epithelial-mesenchymal transition. Biomed Res Int. 2014; 2014:530786.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The MicroRNA hsa-let-7g Promotes Proliferation and Inhibits Apoptosis in Lung Cancer by Targeting HOXB1
- Antiproliferative effect of Citrus junos extracts on A549 human non-smallcell lung cancer cells
- The Incidence Rate and Severity of Orthotopic Lung Cancer in an Animal Model Depends on the Number of A549 Cells and Transplantation Period
- Sanghuangporus sanghuang extract inhibits the proliferation and invasion of lung cancer cells in vitro and in vivo
- Up-Regulation of MiR-1915 Inhibits Proliferation, Invasion, and Migration of Helicobacter pylori-Infected Gastric Cancer Cells via Targeting RAGE