J Vet Sci.
2018 Jul;19(4):528-535. 10.4142/jvs.2018.19.4.528.
Identification of determinants that mediate binding between Tembusu virus and the cellular receptor heat shock protein A9
- Affiliations
-
- 1Institute of Veterinary Medicine, Jiangsu Academy of Agricultural Sciences, Key Laboratory of Veterinary Biological Engineering and Technology, Ministry of Agriculture, Nanjing 210014, China. zhaodongmin126@126.com
- KMID: 2417568
- DOI: http://doi.org/10.4142/jvs.2018.19.4.528
Abstract
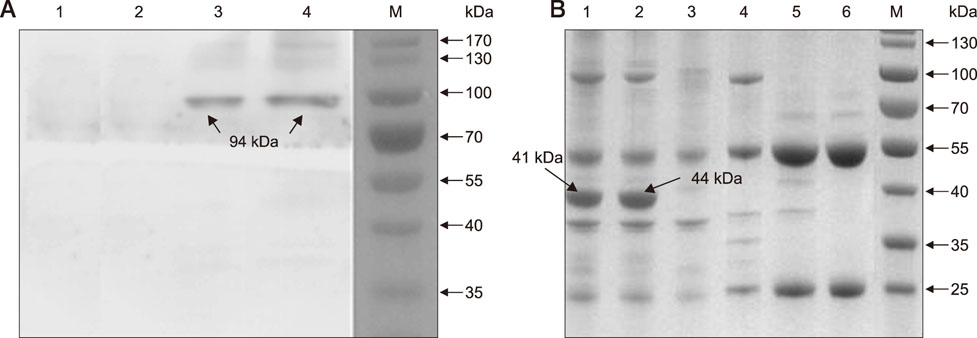
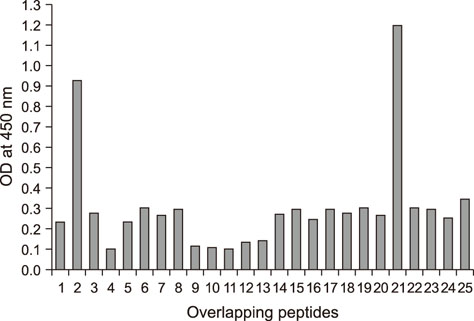
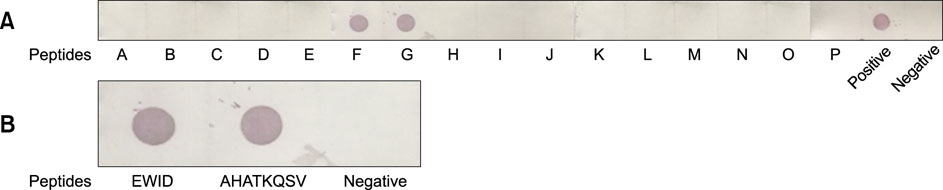
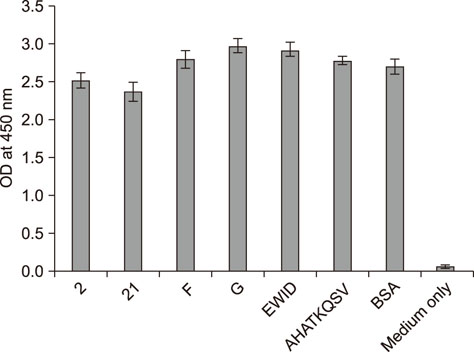
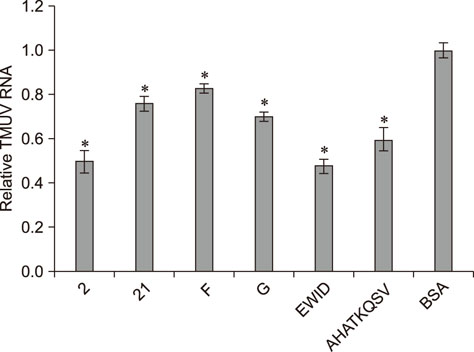
- Heat shock protein A9 (HSPA9), a member of the heat shock protein family, is a putative receptor for Tembusu virus (TMUV). By using Western blot and co-immunoprecipitation assays, E protein domains I and II were identified as the functional domains that facilitate HSPA9 binding. Twenty-five overlapping peptides covering domain I and domain II sequences were synthesized and analyzed by using an HSPA9 binding assay. Two peptides showed the capability of binding to HSPA9. Dot blot assay of truncated peptides indicated that amino acid residues 19 to 22 and 245 to 252 of E protein constitute the minimal motifs required for TMUV binding to HSPA9. Importantly, peptides harboring those two minimal motifs could effectively inhibit TMUV infection. Our results provide insight into TMUV-receptor interaction, thereby creating opportunities for elucidating the mechanism of TMUV entry.
MeSH Terms
Figure
Reference
-
1. Allison SL, Schalich J, Stiasny K, Mandl CW, Heinz FX. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J Virol. 2001; 75:4268–4275.
Article2. Brault JB, Kudelko M, Vidalain PO, Tangy F, Desprès P, Pardigon N. The interaction of flavivirus M protein with light chain Tctex-1 of human dynein plays a role in late stages of virus replication. Virology. 2011; 417:369–378.
Article3. Bressanelli S, Stiasny K, Allison SL, Stura EA, Duquerroy S, Lescar J, Heinz FX, Rey FA. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 2004; 23:728–738.
Article4. Chanel-Vos C, Kielian M. A conserved histidine in the ij loop of the Semliki Forest virus E1 protein plays an important role in membrane fusion. J Virol. 2004; 78:13543–13552.
Article5. Chu JJ, Rajamanonmani R, Li J, Bhuvanakantham R, Lescar J, Ng ML. Inhibition of West Nile virus entry by using a recombinant domain III from the envelope glycoprotein. J Gen Virol. 2005; 86:405–412.
Article6. Dai L, Li Z, Tao P. Evolutionary analysis of Tembusu virus: evidence for the emergence of a dominant genotype. Infect Genet Evol. 2015; 32:124–129.
Article7. Das S, Laxminarayana SV, Chandra N, Ravi V, Desai A. Heat shock protein 70 on Neuro2a cells is a putative receptor for Japanese encephalitis virus. Virology. 2009; 385:47–57.
Article8. Han K, Zhao D, Liu Y, Liu Q, Huang X, Yang J, Bi K, Xu T, Li Y. Generation and characterization of a monoclonal antibody against duck Tembusu virus envelope protein. Pol J Vet Sci. 2016; 19:877–883.
Article9. Harrison SC. The pH sensor for flavivirus membrane fusion. J Cell Biol. 2008; 183:177–179.
Article10. He Y, Wang A, Chen S, Wu Z, Zhang J, Wang M, Jia R, Zhu D, Liu M, Yang Q, Wu Y, Sun K, Chen X, Cheng A. Differential immune-related gene expression in the spleens of duck Tembusu virus-infected goslings. Vet Microbiol. 2017; 212:39–47.
Article11. Huang X, Han K, Zhao D, Liu Y, Zhang J, Niu H, Zhang K, Zhu J, Wu D, Gao L, Li Y. Identification and molecular characterization of a novel flavivirus isolated from geese in China. Res Vet Sci. 2013; 94:774–780.
Article12. Lee JW, Chu JJ, Ng ML. Quantifying the specific binding between West Nile virus envelope domain III protein and the cellular receptor αVβ3 integrin. J Biol Chem. 2006; 281:1352–1360.
Article13. Li C, Zhang LY, Sun MX, Li PP, Huang L, Wei JC, Yao YL, Isahg H, Chen PY, Mao X. Inhibition of Japanese encephalitis virus entry into the cells by the envelope glycoprotein domain III (EDIII) and the loop3 peptide derived from EDIII. Antiviral Res. 2012; 94:179–183.
Article14. Li S, Li X, Zhang L, Wang Y, Yu X, Tian K, Su W, Han B, Su J. Duck Tembusu virus exhibits neurovirulence in BALB/c mice. Virol J. 2013; 10:260.
Article15. Liang JJ, Yu CY, Liao CL, Lin YL. Vimentin binding is critical for infection by the virulent strain of Japanese encephalitis virus. Cell Microbiol. 2011; 13:1358–1370.
Article16. Lin TW, Lo CW, Lai SY, Fan RJ, Lo CJ, Chou YM, Thiruvengadam R, Wang AH, Wang MY. Chicken heat shock protein 90 is a component of the putative cellular receptor complex of infectious bursal disease virus. J Virol. 2007; 81:8730–8741.
Article17. Liu H, Liu Y, Wang S, Zhang Y, Zu X, Zhou Z, Zhang B, Xiao G. Structure-based mutational analysis of several sites in the E protein: implications for understanding the entry mechanism of Japanese encephalitis virus. J Virol. 2015; 89:5668–5686.
Article18. Liu M, Chen S, Chen Y, Liu C, Chen S, Yin X, Li G, Zhang Y. Adapted Tembusu-like virus in chickens and geese in China. J Clin Microbiol. 2012; 50:2807–2809.
Article19. Liu Q, Huang X, Zhao D, Han K, Liu Y, Yang J, Bi K, Li Y. Identification of heat shock protein A9 as a Tembusu virus binding protein on DF-1 cells. Virus Res. 2017; 227:110–114.
Article20. Medigeshi GR, Hirsch AJ, Streblow DN, Nikolich-Zugich J, Nelson JA. West Nile virus entry requires cholesterol-rich membrane microdomains and is independent of αvβ3 integrin. J Virol. 2008; 82:5212–5219.
Article21. Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004; 427:313–319.
Article22. Modis Y, Ogata S, Clements D, Harrison SC. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J Virol. 2005; 79:1223–1231.
Article23. Nelson S, Poddar S, Lin TY, Pierson TC. Protonation of individual histidine residues is not required for the pH-dependent entry of West Nile virus: evaluation of the “histidine switch” hypothesis. J Virol. 2009; 83:12631–12635.
Article24. Oliphant T, Nybakken GE, Engle M, Xu Q, Nelson CA, Sukupolvi-Petty S, Marri A, Lachmi BE, Olshevsky U, Fremont DH, Pierson TC, Diamond MS. Antibody recognition and neutralization determinants on domains I and II of West Nile virus envelope protein. J Virol. 2006; 80:12149–12159.
Article25. Pokidysheva E, Zhang Y, Battisti AJ, Bator-Kelly CM, Chipman PR, Xiao C, Gregorio GG, Hendrickson WA, Kuhn RJ, Rossmann MG. Cryo-EM reconstruction of dengue virus in complex with the carbohydrate recognition domain of DC-SIGN. Cell. 2006; 124:485–493.
Article26. Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. Am J Hyg. 1938; 27:493–497.27. Reyes-Del Valle J, Chávez-Salinas S, Medina F, Del Angel RM. Heat shock protein 90 and heat shock protein 70 are components of dengue virus receptor complex in human cells. J Virol. 2005; 79:4557–4567.
Article28. Rodenhuis-Zybert IA, Wilschut J, Smit JM. Dengue virus life cycle: viral and host factors modulating infectivity. Cell Mol Life Sci. 2010; 67:2773–2786.
Article29. Tang Y, Diao Y, Chen H, Ou Q, Liu X, Gao X, Yu C, Wang L. Isolation and genetic characterization of a Tembusu virus strain isolated from mosquitoes in Shandong, China. Transbound Emerg Dis. 2015; 62:209–216.
Article30. Tang Y, Diao Y, Yu C, Gao X, Ju X, Xue C, Liu X, Ge P, Qu J, Zhang D. Characterization of a Tembusu virus isolated from naturally infected house sparrows (Passer domesticus) in Northern China. Transbound Emerg Dis. 2013; 60:152–158.
Article31. Tang Y, Gao X, Diao Y, Feng Q, Chen H, Liu X, Ge P, Yu C. Tembusu virus in human, China. Transbound Emerg Dis. 2013; 60:193–196.
Article32. Thepparit C, Smith DR. Serotype-specific entry of dengue virus into liver cells: identification of the 37-kilodalton/67-kilodalton high-affinity laminin receptor as a dengue virus serotype 1 receptor. J Virol. 2004; 78:12647–12656.
Article33. Ti J, Zhang M, Li Z, Li X, Diao Y. Duck Tembusu virus exhibits pathogenicity to Kunming mice by intracerebral inoculation. Front Microbiol. 2016; 7:190.
Article34. Wang HJ, Li XF, Liu L, Xu YP, Ye Q, Deng YQ, Huang XY, Zhao H, Qin ED, Shi PY, Gao GF, Qin CF. The emerging duck flavivirus is not pathogenic for primates and is highly sensitive to mammalian interferon antiviral signaling. J Virol. 2016; 90:6538–6548.
Article35. Wang P, Hu K, Luo S, Zhang M, Deng X, Li C, Jin W, Hu B, He S, Li M, Du T, Xiao G, Zhang B, Liu Y, Hu Q. DC-SIGN as an attachment factor mediates Japanese encephalitis virus infection of human dendritic cells via interaction with a single high-mannose residue of viral E glycoprotein. Virology. 2016; 488:108–119.
Article36. Wang Y, Yuan X, Li Y, Yu K, Yang J, Xu H, Zhang Y, Yu K, Liao M, Qin Z. Rapid detection of newly isolated Tembusu-related Flavivirus by reverse-transcription loop-mediated isothermal amplification assay. Virol J. 2011; 8:553.
Article37. Wei Y, Ma Y, Luo L, Wu X, Huang Y, Li X, Yang Z. Differences in clinical and laboratory features for different genotypes of Orientia tsutsugamushi in Guangzhou, Southern China. Vector Borne Zoonotic Dis. 2017; 17:260–267.
Article38. Yu K, Sheng ZZ, Huang B, Ma X, Li Y, Yuan X, Qin Z, Wang D, Chakravarty S, Li F, Song M, Sun H. Structural, antigenic, and evolutionary characterizations of the envelope protein of newly emerging Duck Tembusu Virus. PLoS One. 2013; 8:e71319.
Article39. Zhao D, Huang X, Liu Y, Han K, Zhang J, Yang J, Xie X, Li Y. Domain I and II from newly emerging goose tembusu virus envelope protein functions as a dominant-negative inhibitor of virus infectivity. Res Vet Sci. 2015; 98:121–126.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Expression of Heat Shock Protein 70 m-RNA in Rat Bladder Overdistended by Diuresis
- Environmental factors regulating the expression of Porphyromonas gingivalis heat shock protein
- Heat Shock Protein 90 (HSP90) and Immune Regulation
- Identification of a putative cellular receptor 150 kDa polypeptide for porcine epidemic diarrhea virus in porcine enterocytes
- Identification of a Major Cytoplasmic Laminin Binding Protein as the Heat Shock Cognate Protein 70 in Highly Metastatic Human Colon Carcinoma Cells