Korean J Physiol Pharmacol.
2018 Jul;22(4):399-408. 10.4196/kjpp.2018.22.4.399.
Lipidomic analysis of plasma lipids composition changes in septic mice
- Affiliations
-
- 1Department of Pharmacology, College of Medicine, Institute of Natural Medicine, Hallym University, Chuncheon 24252, Korea. dksong@hallym.ac.kr
- KMID: 2414264
- DOI: http://doi.org/10.4196/kjpp.2018.22.4.399
Abstract
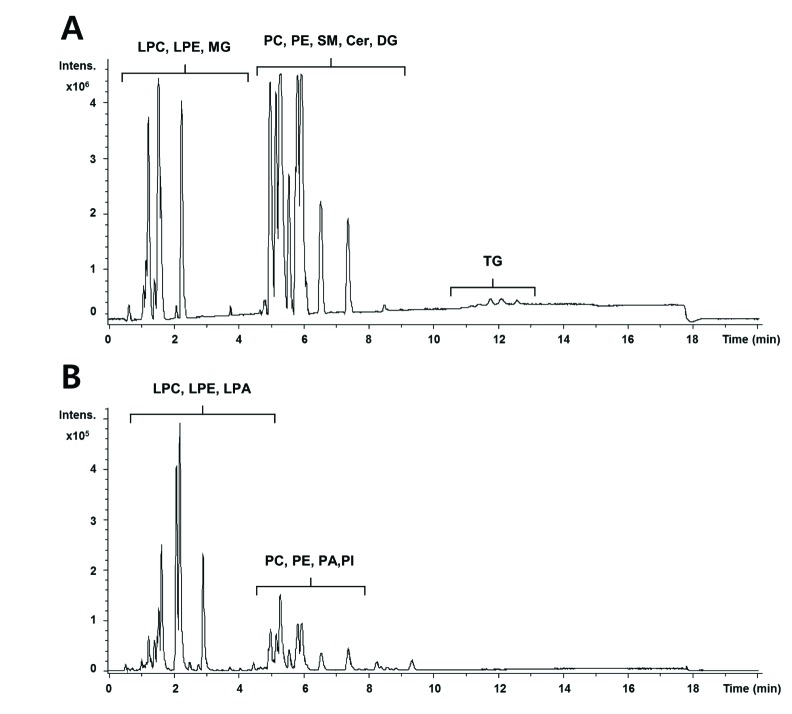
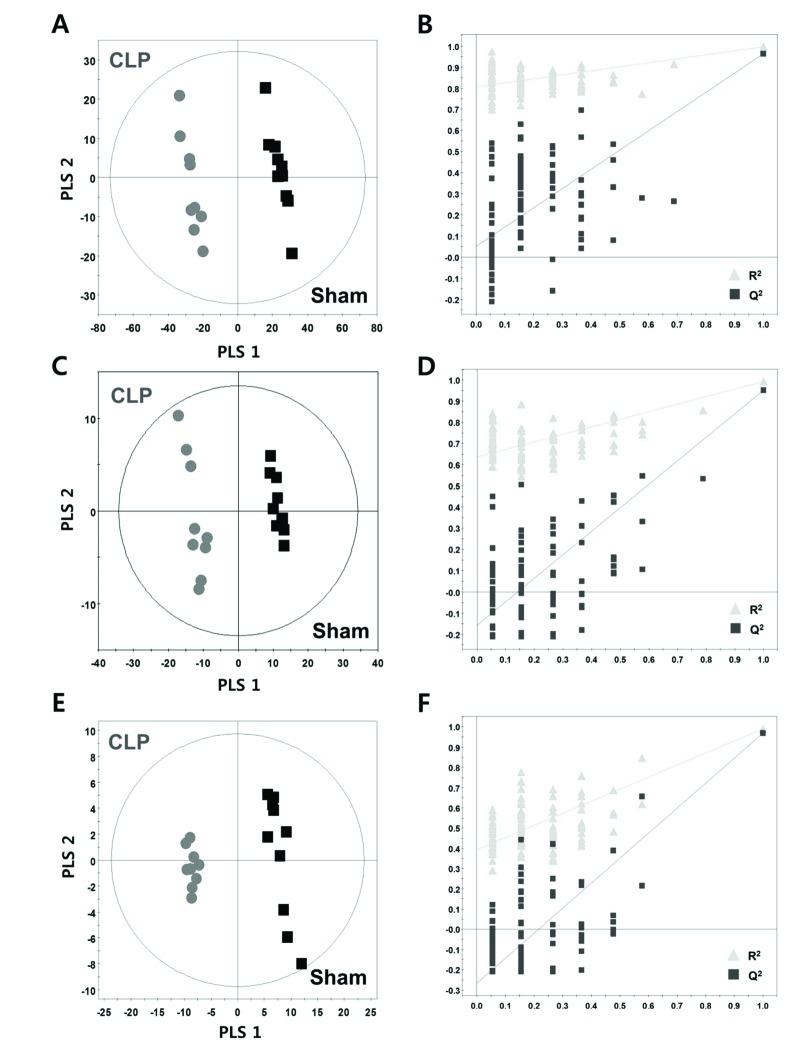
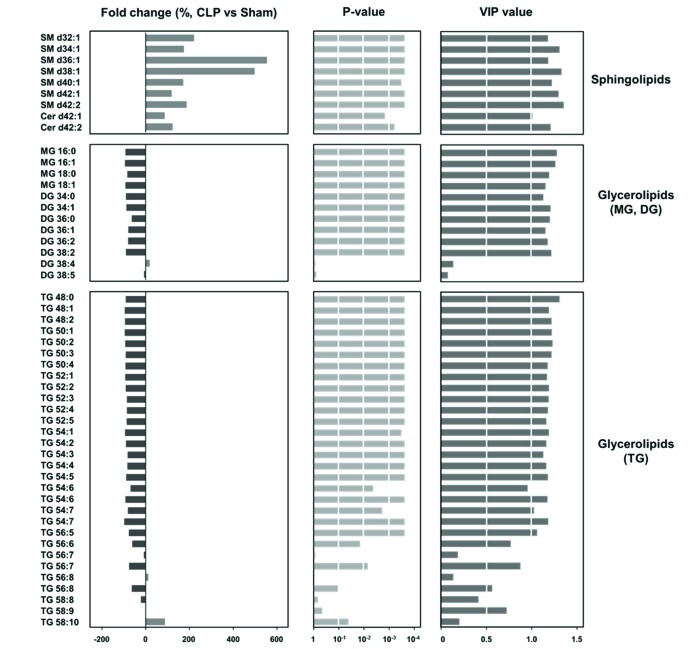
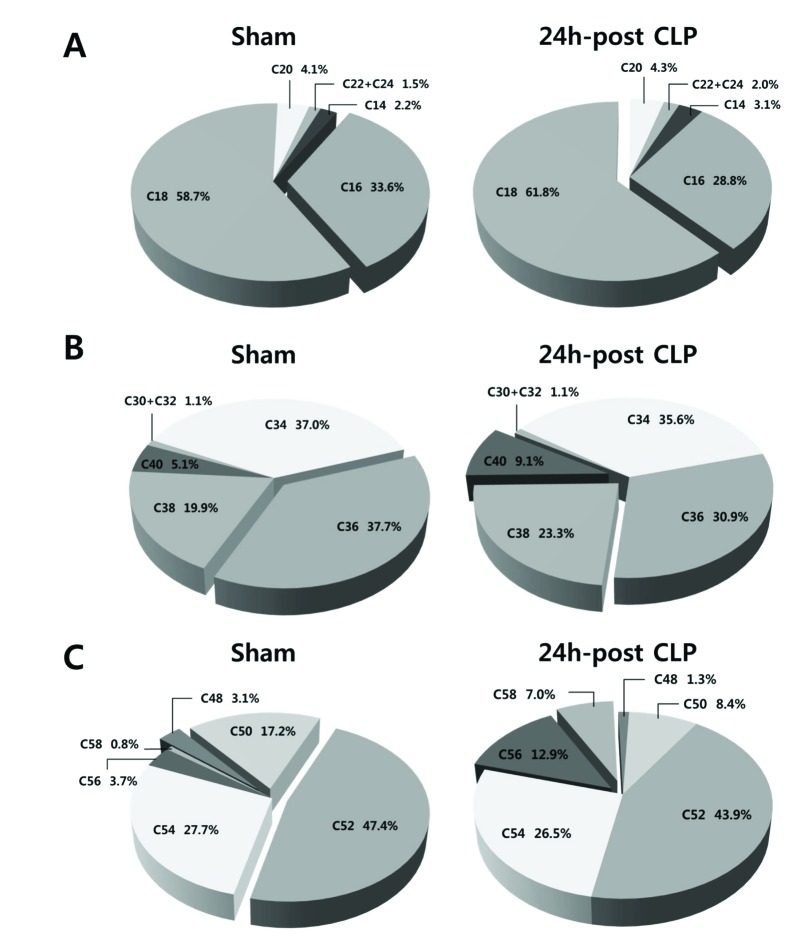
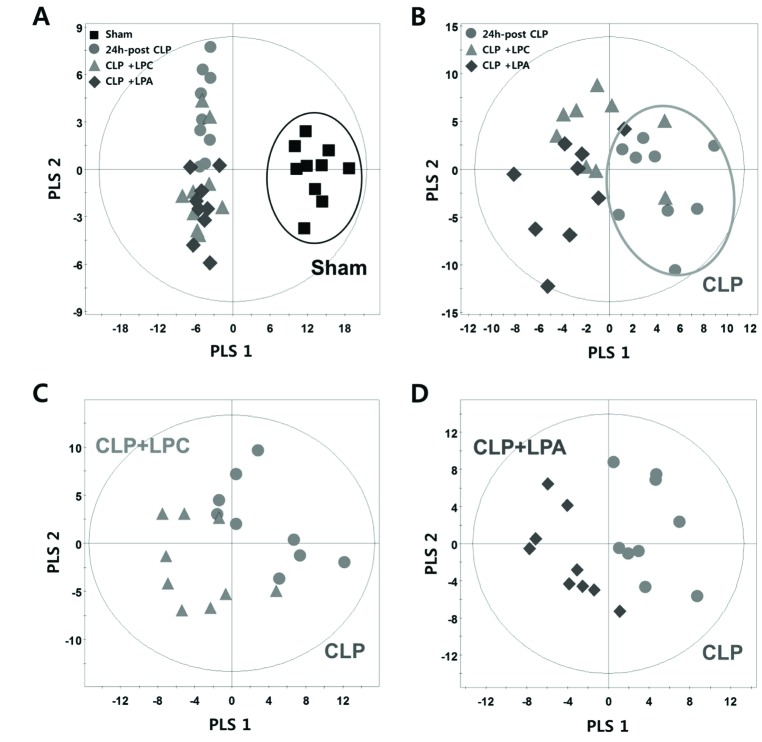
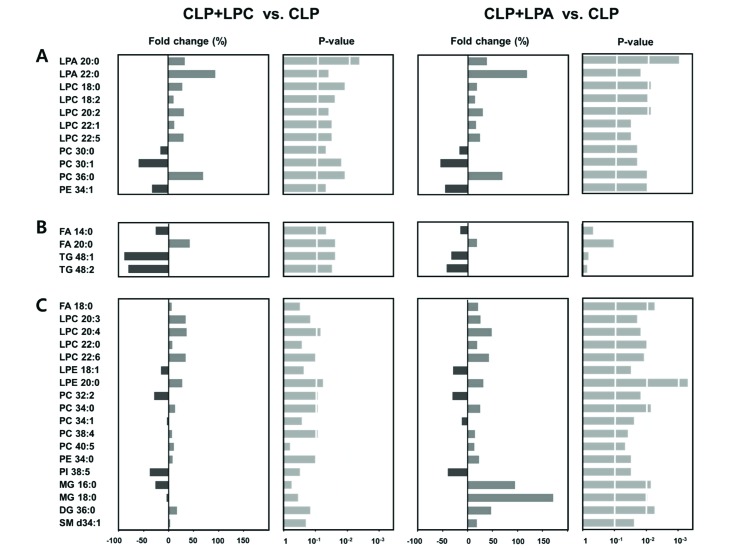
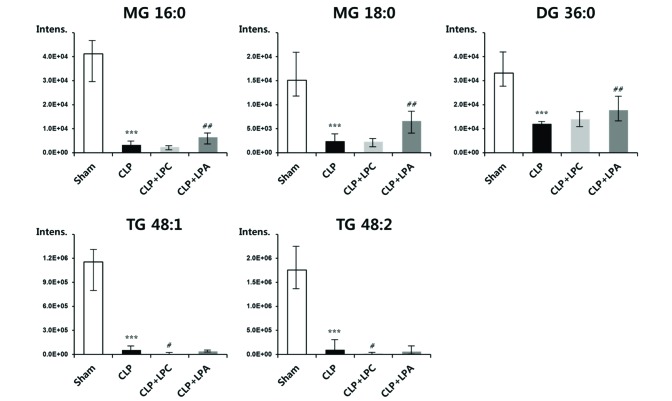
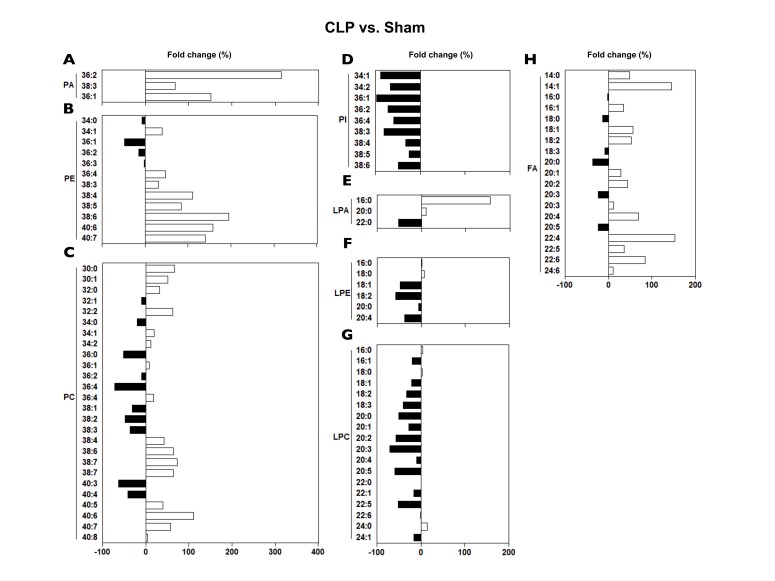
- A lipidomic study on extensive plasma lipids in bacterial peritonitis (cecal ligation and puncture, CLP)-induced sepsis in mice was done at 24 h post-CLP. The effects of administration of lysophosphatidylcholine (LPC) and lysophosphatidic acid (LPA), compounds known to have beneficial effects in CLP, on the sepsis-induced plasma lipid changes were also examined. Among the 147 plasma lipid species from 13 lipid subgroups (fatty acid [FA], LPA, LPC, lysophosphatidylethanolamine [LPE], phosphatidic acid [PA], phosphatidylcholine [PC], phosphatidylethanolamine [PE], phosphatidylinositol [PI], monoacylglyceride [MG], diacylglyceride [DG], triacylglyceride [TG], sphingomyelin [SM], and ceramide [Cer]) analyzed in this study, 40 and 70 species were increased, and decreased, respectively, in the CLP mice. Treatments with LPC and LPA affected 14 species from 7 subgroups, and 25 species from 9 subgroups, respectively. These results could contribute to finding the much needed reliable biomarkers of sepsis.
Keyword
MeSH Terms
Figure
Reference
-
1. Ghosh A, Nishtala K. Biofluid lipidome: a source for potential diagnostic biomarkers. Clin Transl Med. 2017; 6:22. PMID: 28639235.
Article2. Quehenberger O, Armando AM, Brown AH, Milne SB, Myers DS, Merrill AH, Bandyopadhyay S, Jones KN, Kelly S, Shaner RL, Sullards CM, Wang E, Murphy RC, Barkley RM, Leiker TJ, Raetz CR, Guan Z, Laird GM, Six DA, Russell DW, McDonald JG, Subramaniam S, Fahy E, Dennis EA. Lipidomics reveals a remarkable diversity of lipids in human plasma. J Lipid Res. 2010; 51:3299–3305. PMID: 20671299.
Article3. Drobnik W, Liebisch G, Audebert FX, Frohlich D, Gluck T, Vogel P, Rothe G, Schmitz G. Plasma ceramide and lysophosphatidylcholine inversely correlate with mortality in sepsis patients. J Lipid Res. 2003; 44:754–761. PMID: 12562829.
Article4. Levels JH, Pajkrt D, Schultz M, Hoek FJ, van Tol A, Meijers JC, van Deventer SJ. Alterations in lipoprotein homeostasis during human experimental endotoxemia and clinical sepsis. Biochim Biophys Acta. 2007; 1771:1429–1438. PMID: 17980169.
Article5. Nogueira AC, Kawabata V, Biselli P, Lins MH, Valeri C, Seckler M, Hoshino W, Júnior LG, Bernik MM, de Andrade Machado JB, Martinez MB, Lotufo PA, Caldini EG, Martins E, Curi R, Soriano FG. Changes in plasma free fatty acid levels in septic patients are associated with cardiac damage and reduction in heart rate variability. Shock. 2008; 29:342–348. PMID: 18000476.
Article6. Schmerler D, Neugebauer S, Ludewig K, Bremer-Streck S, Brunkhorst FM, Kiehntopf M. Targeted metabolomics for discrimination of systemic inflammatory disorders in critically ill patients. J Lipid Res. 2012; 53:1369–1375. PMID: 22581935.
Article7. Rival T, Cinq-Frais C, Silva-Sifontes S, Garcia J, Riu B, Salvayre R, Genestal M, Caspar-Bauguil S. Alteration of plasma phospholipid fatty acid profile in patients with septic shock. Biochimie. 2013; 95:2177–2181. PMID: 23954620.
Article8. Park DW, Kwak DS, Park YY, Chang Y, Huh JW, Lim CM, Koh Y, Song DK, Hong SB. Impact of serial measurements of lysophosphatidylcholine on 28-day mortality prediction in patients admitted to the intensive care unit with severe sepsis or septic shock. J Crit Care. 2014; 29:882882.e5–882.e11.
Article9. Rogers AJ, McGeachie M, Baron RM, Gazourian L, Haspel JA, Nakahira K, Fredenburgh LE, Hunninghake GM, Raby BA, Matthay MA, Otero RM, Fowler VG, Rivers EP, Woods CW, Kingsmore S, Langley RJ, Choi AM. Metabolomic derangements are associated with mortality in critically ill adult patients. PLoS One. 2014; 9:e87538. PMID: 24498130.
Article10. Ferrario M, Cambiaghi A, Brunelli L, Giordano S, Caironi P, Guatteri L, Raimondi F, Gattinoni L, Latini R, Masson S, Ristagno G, Pastorelli R. Mortality prediction in patients with severe septic shock: a pilot study using a target metabolomics approach. Sci Rep. 2016; 6:20391. PMID: 26847922.
Article11. Bermudes ACG, de Carvalho WB, Zamberlan P, Muramoto G, Maranhão RC, Delgado AF. Changes in lipid metabolism in pediatric patients with severe sepsis and septic shock. Nutrition. 2018; 47:104–109. PMID: 29429528.
Article12. Xu PB, Lin ZY, Meng HB, Yan SK, Yang Y, Liu XR, Li JB, Deng XM, Zhang WD. A metabonomic approach to early prognostic evaluation of experimental sepsis. J Infect. 2008; 56:474–481. PMID: 18471887.
Article13. Rittirsch D, Huber-Lang MS, Flierl MA, Ward PA. Immunodesign of experimental sepsis by cecal ligation and puncture. Nat Protoc. 2009; 4:31–36. PMID: 19131954.
Article14. Yan JJ, Jung JS, Lee JE, Lee J, Huh SO, Kim HS, Jung KC, Cho JY, Nam JS, Suh HW, Kim YH, Song DK. Therapeutic effects of lysophosphatidylcholine in experimental sepsis. Nat Med. 2004; 10:161–167. PMID: 14716308.
Article15. Ahn WG, Jung JS, Kwon HY, Song DK. Alteration of lysophosphatidylcholine-related metabolic parameters in the plasma of mice with experimental sepsis. Inflammation. 2017; 40:537–545. PMID: 28028754.
Article16. Folch J, Lees M, Sloane-Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957; 226:497–509. PMID: 13428781.
Article17. Wong ML, Xie B, Beatini N, Phu P, Marathe S, Johns A, Gold PW, Hirsch E, Williams KJ, Licinio J, Tabas I. Acute systemic inflammation up-regulates secretory sphingomyelinase in vivo: a possible link between inflammatory cytokines and atherogenesis. Proc Natl Acad Sci U S A. 2000; 97:8681–8686. PMID: 10890909.
Article18. Rice GC, Brown PA, Nelson RJ, Bianco JA, Singer JW, Bursten S. Protection from endotoxic shock in mice by pharmacologic inhibition of phosphatidic acid. Proc Natl Acad Sci U S A. 1994; 91:3857–3861. PMID: 8171002.
Article19. Sokolović M, Sokolović A, van Roomen CP, Gruber A, Ottenhoff R, Scheij S, Hakvoort TB, Lamers WH, Groen AK. Unexpected effects of fasting on murine lipid homeostasis−transcriptomic and lipid profiling. J Hepatol. 2010; 52:737–744. PMID: 20347175.20. Pierrakos C, Vincent JL. Sepsis biomarkers: a review. Crit Care. 2010; 14:R15. PMID: 20144219.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Effect of Combined Training at Different Times of Day on Body Composition, Plasma Lipids, Stress Hormones and Nutrient Intakes
- Lipids Analysis of Epidermis and Stratum corneum Using Circumcised Prepuce
- Quantitative Analysis of Solvent Extracted Skin Surface Lipid in Human Skin ( - hexane, hexane/ methanol, ethanol - )
- Blueberry, blackberry, and blackcurrant differentially affect plasma lipids and pro-inflammatory markers in diet-induced obesity mice
- Dietary Oxidized Linoleic Acids Modulate Fatty Acids in Mice