J Breast Cancer.
2018 Jun;21(2):142-149. 10.4048/jbc.2018.21.2.142.
Combined Let-7a and H19 Signature: A Prognostic Index of Progression-Free Survival in Primary Breast Cancer Patients
- Affiliations
-
- 1Guangdong Provincial Key Laboratory of Malignant Tumor Epigenetics and Gene Regulation, Guangzhou, China. fengyanyu_summer@163.com
- 2Department of Breast Surgery, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, China.
- 3Department of Medical Oncology, Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education), Peking University Cancer Hospital and Institute, Peking, China.
- KMID: 2413932
- DOI: http://doi.org/10.4048/jbc.2018.21.2.142
Abstract
- PURPOSE
The long non-coding RNA H19, a conservatively imprinted gene, acts as a molecular sponge for the let-7 family, which has been identified as a set of tumor suppressors. However, the combined prognostic value of H19 and let-7a signature in breast cancer patients remains unclear.
METHODS
In this research we assessed the prognostic value of the combined H19 and let-7a signature in breast cancer patients by retrospectively reviewing that data of 79 patients who underwent neoadjuvant chemotherapy; we also investigated the expression and function of H19 in breast cancer cell lines in vitro. Survival data were calculated using the Kaplan-Meier method and compared using the log-rank test. Univariate and multivariate survival analyses were conducted using the Cox proportional hazards regression method. As determined using X-tile, the optimal cutoff value for the risk score to assess progression-free survival (PFS) based on the combined signature was -0.1.
RESULTS
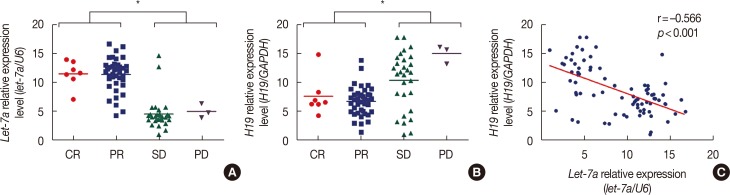
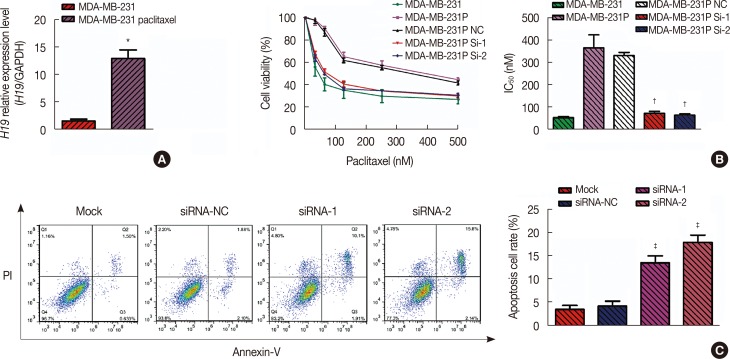
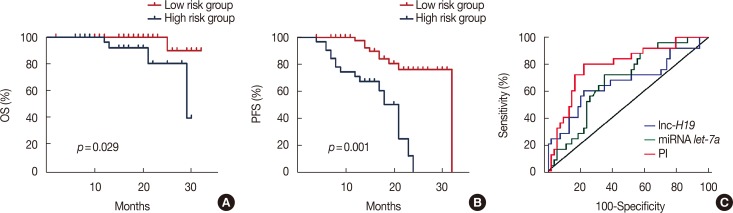
Patients with an overall positive treatment response had higher let-7a and lower H19 levels. In addition, let-7a expression was negatively correlated with H19 expression. Patients with a risk score of >-0.1 had shorter overall survival and PFS. In vitro data showed that chemoresistant cell lines exhibit higher H19 and lower let-7a levels and knockdown H19 restores paclitaxel sensitivity.
CONCLUSION
Our results suggest that the combined let-7a and H19 signature is a novel prognostic factor for breast cancer patients treated with neoadjuvant chemotherapy.
Keyword
MeSH Terms
Figure
Reference
-
1. Forouzanfar MH, Foreman KJ, Delossantos AM, Lozano R, Lopez AD, Murray CJ, et al. Breast and cervical cancer in 187 countries between 1980 and 2010: a systematic analysis. Lancet. 2011; 378:1461–1484. PMID: 21924486.
Article2. Kaufmann M, von Minckwitz G, Bear HD, Buzdar A, McGale P, Bonnefoi H, et al. Recommendations from an international expert panel on the use of neoadjuvant (primary) systemic treatment of operable breast cancer: new perspectives 2006. Ann Oncol. 2007; 18:1927–1934. PMID: 17998286.
Article3. Thum T, Condorelli G. Long noncoding RNAs and microRNAs in cardiovascular pathophysiology. Circ Res. 2015; 116:751–762. PMID: 25677521.
Article4. Voortman J, Goto A, Mendiboure J, Sohn JJ, Schetter AJ, Saito M, et al. MicroRNA expression and clinical outcomes in patients treated with adjuvant chemotherapy after complete resection of non-small cell lung carcinoma. Cancer Res. 2010; 70:8288–8298. PMID: 20978195.
Article5. He FC, Meng WW, Qu YH, Zhou MX, He J, Lv P, et al. Expression of circulating microRNA-20a and let-7a in esophageal squamous cell carcinoma. World J Gastroenterol. 2015; 21:4660–4665. PMID: 25914476.
Article6. Vennin C, Spruyt N, Dahmani F, Julien S, Bertucci F, Finetti P, et al. H19 non coding RNA-derived miR-675 enhances tumorigenesis and metastasis of breast cancer cells by downregulating c-Cbl and Cbl-b. Oncotarget. 2015; 6:29209–29223. PMID: 26353930.7. Si X, Zang R, Zhang E, Liu Y, Shi X, Zhang E, et al. LncRNA H19 confers chemoresistance in ERα-positive breast cancer through epigenetic silencing of the pro-apoptotic gene BIK. Oncotarget. 2016; 7:81452–81462. PMID: 27845892.
Article8. Peng F, Li TT, Wang KL, Xiao GQ, Wang JH, Zhao HD, et al. H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis. 2017; 8:e2569. PMID: 28102845.
Article9. Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013; 52:101–112. PMID: 24055342.
Article10. Wu J, Li S, Jia W, Deng H, Chen K, Zhu L, et al. Reduced let-7a is associated with chemoresistance in primary breast cancer. PLoS One. 2015; 10:e0133643. PMID: 26218285.
Article11. Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004; 10:7252–7259. PMID: 15534099.12. Rastogi P, Anderson SJ, Bear HD, Geyer CE, Kahlenberg MS, Robidoux A, et al. Preoperative chemotherapy: updates of National Surgical Adjuvant Breast and Bowel Project Protocols B-18 and B-27. J Clin Oncol. 2008; 26:778–785. PMID: 18258986.
Article13. Mauri D, Pavlidis N, Ioannidis JP. Neoadjuvant versus adjuvant systemic treatment in breast cancer: a meta-analysis. J Natl Cancer Inst. 2005; 97:188–194. PMID: 15687361.
Article14. Gabory A, Jammes H, Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays. 2010; 32:473–480. PMID: 20486133.
Article15. Onyango P, Feinberg AP. A nucleolar protein, H19 opposite tumor suppressor (HOTS), is a tumor growth inhibitor encoded by a human imprinted H19 antisense transcript. Proc Natl Acad Sci U S A. 2011; 108:16759–16764. PMID: 21940503.
Article16. Li S, Hua Y, Jin J, Wang H, Du M, Zhu L, et al. Association of genetic variants in lncRNA H19 with risk of colorectal cancer in a Chinese population. Oncotarget. 2016; 7:25470–25477. PMID: 27027436.
Article17. Zhou X, Yin C, Dang Y, Ye F, Zhang G. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep. 2015; 5:11516. PMID: 26096073.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The 70-Gene Prognostic Signature for Korean Breast Cancer Patients
- Prognostic factors in breast cancer with extracranial oligometastases and the appropriate role of radiation therapy
- The Prognostic Significance of p27 and cyclin E in Human Breast Cancer
- Molecular Signature for Lymphatic Invasion Associated with Survival of Epithelial Ovarian Cancer
- Outcome of Surgical Excision for Isolated Locoregional Recurrence of Breast Cancer