J Breast Cancer.
2018 Jun;21(2):112-123. 10.4048/jbc.2018.21.2.112.
Histone Deacetylase-3 Modification of MicroRNA-31 Promotes Cell Proliferation and Aerobic Glycolysis in Breast Cancer and Is Predictive of Poor Prognosis
- Affiliations
-
- 1Department of Pathology, Suining Central Hospital, Suining, China. zhyf3270@163.com
- KMID: 2413929
- DOI: http://doi.org/10.4048/jbc.2018.21.2.112
Abstract
- PURPOSE
The incidence and mortality of breast cancer is increasing worldwide. There is a constant quest to understand the underlying molecular biology of breast cancer so as to plan better treatment options. The purpose of the current study was to characterize the expression of histone deacetylases-3 (HDAC3), a member of class I HDACs, and assess the clinical significance of HDAC3 in breast cancer.
METHODS
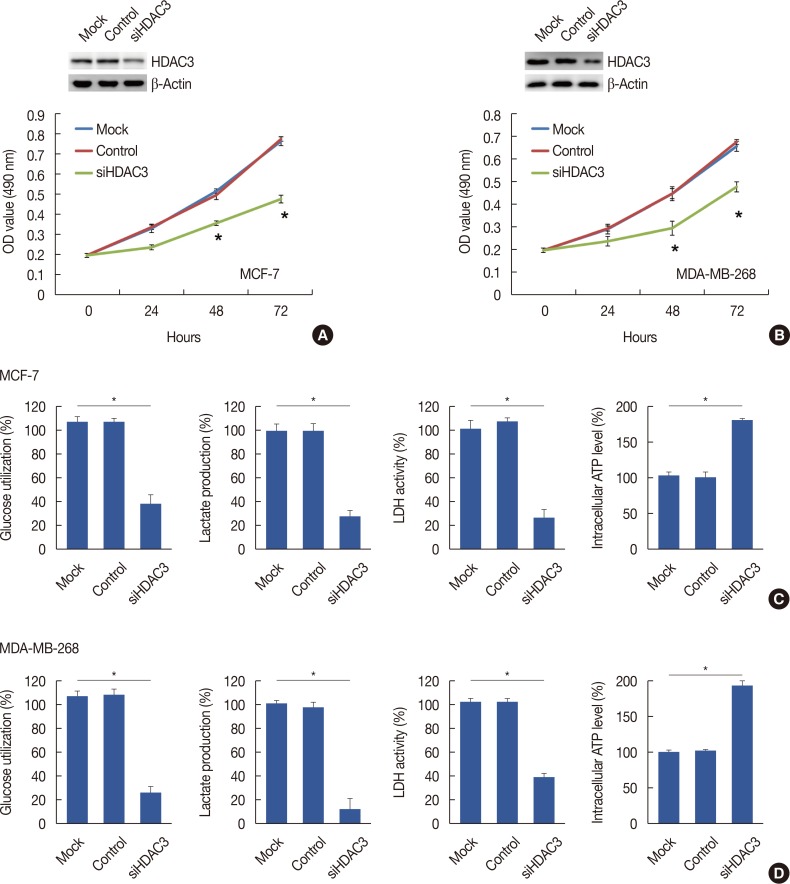
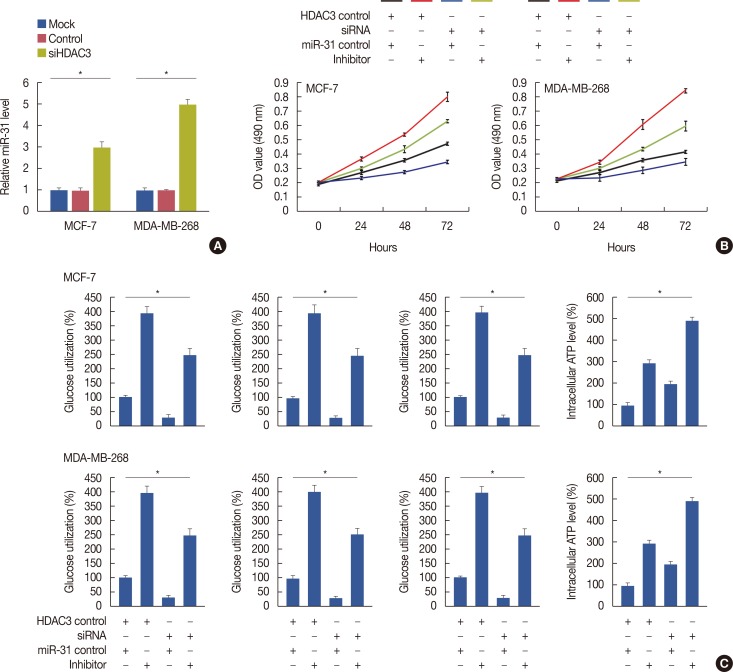
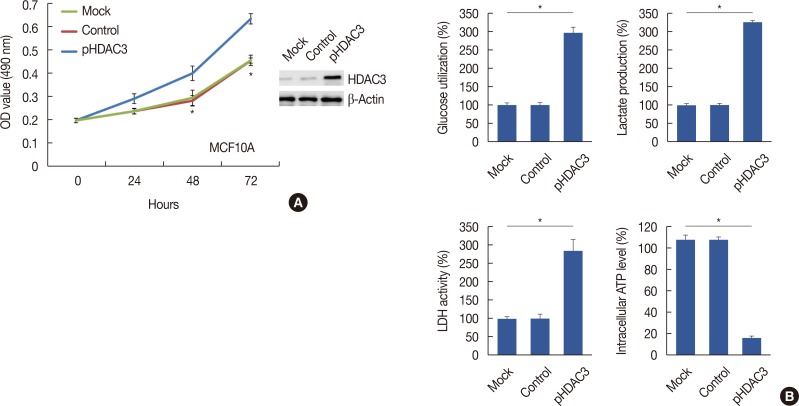
Quantitative real-time polymerase chain reaction, immunohistochemistry, and western blot analysis were used to examine messenger RNA and protein expression levels. The relationships between HDAC3 expression and clinicopathological variables were analyzed. MTT assays were used to detect cell proliferation. Glucose-uptake, lactate, adenosine triphosphate, and lactate dehydrogenase assays were employed to detect aerobic glycolysis. Chromatin immunoprecipitation was used to detect microRNA-31 (miR-31) promoter binding.
RESULTS
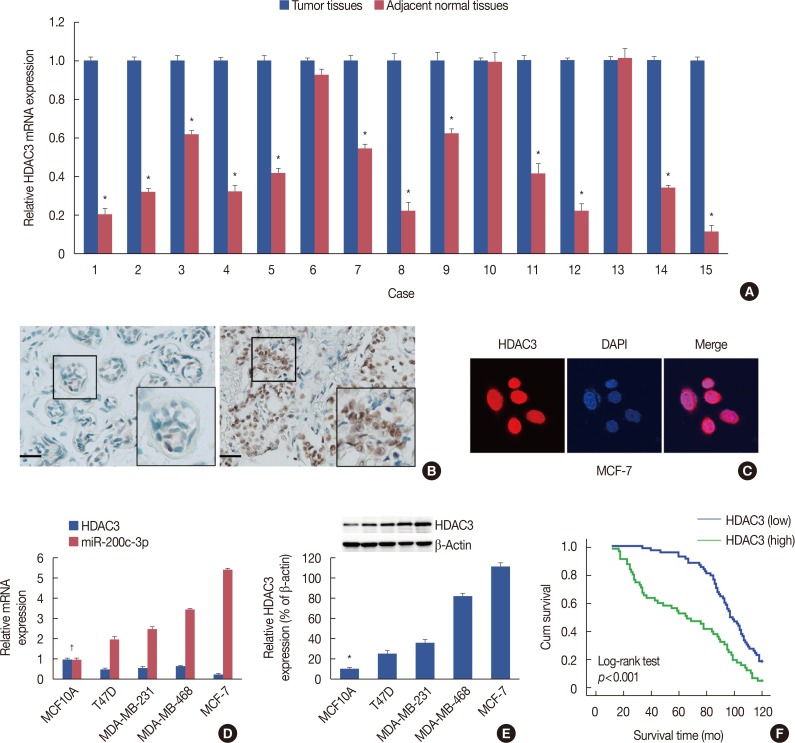
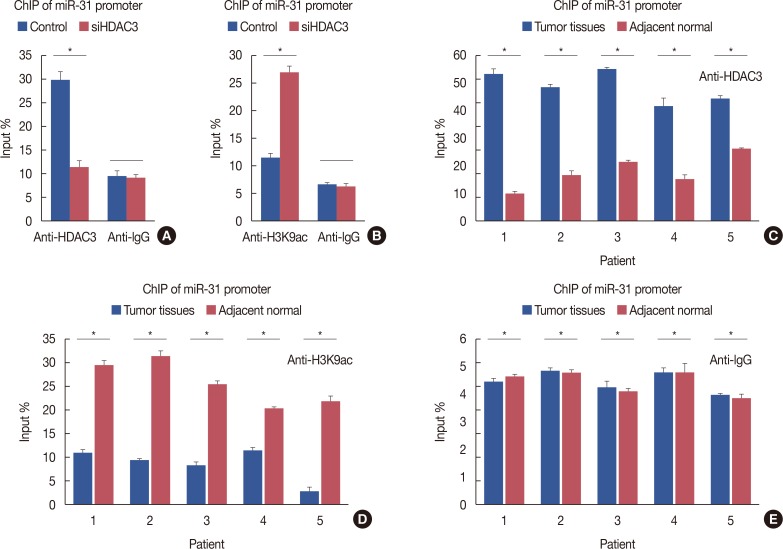
Our data revealed that HDAC3 was upregulated in breast cancer tissue compared with matched para-carcinoma tissues, and high levels of HDAC3 were positively correlated with advanced TNM stage and N stage of cancer. Furthermore, overexpression of HDAC3 promoted breast cancer cell-proliferation and aerobic glycolysis. The functional involvement of HDAC3 was related in part to the repression of miR-31 transcription via decreased histone H3 acetylation at lysine K9 levels of the miR-31 promoter. Survival analysis revealed that the level of HDAC3 was an independent prognostic factor for breast cancer patients.
CONCLUSION
Our findings revealed that HDAC3 served as an oncogene that could promote cell proliferation and aerobic glycolysis and was predictive of a poor prognosis in breast cancer. HDAC3 participated in the cell proliferation of breast cancer, which may prove to be a pivotal epigenetic target against this devastating disease.
MeSH Terms
-
Acetylation
Adenosine Triphosphate
Blotting, Western
Breast Neoplasms*
Breast*
Cell Proliferation*
Chromatin Immunoprecipitation
Epigenomics
Glycolysis*
Histone Code
Histones*
Humans
Immunohistochemistry
Incidence
L-Lactate Dehydrogenase
Lactic Acid
Lysine
Molecular Biology
Mortality
Oncogenes
Prognosis*
Real-Time Polymerase Chain Reaction
Repression, Psychology
RNA, Messenger
Adenosine Triphosphate
Histones
L-Lactate Dehydrogenase
Lactic Acid
Lysine
RNA, Messenger
Figure
Reference
-
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017; 67:7–30. PMID: 28055103.
Article2. Staaf J, Ringnér M, Vallon-Christersson J, Jönsson G, Bendahl PO, Holm K, et al. Identification of subtypes in human epidermal growth factor receptor 2: positive breast cancer reveals a gene signature prognostic of outcome. J Clin Oncol. 2010; 28:1813–1820. PMID: 20231686.3. O'Shaughnessy JA. Molecular signatures predict outcomes of breast cancer. N Engl J Med. 2006; 355:615–617. PMID: 16899783.4. Marmot MG, Altman DG, Cameron DA, Dewar JA, Thompson SG, Wilcox M. The benefits and harms of breast cancer screening: an independent review. Br J Cancer. 2013; 108:2205–2240. PMID: 23744281.
Article5. Kroemer G, Senovilla L, Galluzzi L, André F, Zitvogel L. Natural and therapy-induced immunosurveillance in breast cancer. Nat Med. 2015; 21:1128–1138. PMID: 26444637.
Article6. Brooks MD, Burness ML, Wicha MS. Therapeutic implications of cellular heterogeneity and plasticity in breast cancer. Cell Stem Cell. 2015; 17:260–271. PMID: 26340526.
Article7. Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr Opin Cell Biol. 2001; 13:263–273. PMID: 11343896.
Article8. Buchwald M, Krämer OH, Heinzel T. HDACi: targets beyond chromatin. Cancer Lett. 2009; 280:160–167. PMID: 19342155.9. Legube G, Trouche D. Regulating histone acetyltransferases and deacetylases. EMBO Rep. 2003; 4:944–947. PMID: 14528264.
Article10. Ma Y, Yue Y, Pan M, Sun J, Chu J, Lin X, et al. Histone deacetylase 3 inhibits new tumor suppressor gene DTWD1 in gastric cancer. Am J Cancer Res. 2015; 5:663–673. PMID: 25973305.11. Jiao F, Hu H, Yuan C, Jin Z, Guo Z, Wang L, et al. Histone deacetylase 3 promotes pancreatic cancer cell proliferation, invasion and increases drug-resistance through histone modification of P27, P53 and Bax. Int J Oncol. 2014; 45:1523–1530. PMID: 25070540.
Article12. Pilarsky C, Wenzig M, Specht T, Saeger HD, Grützmann R. Identification and validation of commonly overexpressed genes in solid tumors by comparison of microarray data. Neoplasia. 2004; 6:744–750. PMID: 15720800.
Article13. Müller BM, Jana L, Kasajima A, Lehmann A, Prinzler J, Budczies J, et al. Differential expression of histone deacetylases HDAC1, 2 and 3 in human breast cancer: overexpression of HDAC2 and HDAC3 is associated with clinicopathological indicators of disease progression. BMC Cancer. 2013; 13:215. PMID: 23627572.
Article14. Krusche CA, Wülfing P, Kersting C, Vloet A, Böcker W, Kiesel L, et al. Histone deacetylase-1 and -3 protein expression in human breast cancer: a tissue microarray analysis. Breast Cancer Res Treat. 2005; 90:15–23. PMID: 15770522.
Article15. Seo J, Min SK, Park HR, Kim DH, Kwon MJ, Kim LS, et al. Expression of histone deacetylases HDAC1, HDAC2, HDAC3, and HDAC6 in invasive ductal carcinomas of the breast. J Breast Cancer. 2014; 17:323–331. PMID: 25548579.
Article16. Creighton CJ, Fountain MD, Yu Z, Nagaraja AK, Zhu H, Khan M, et al. Molecular profiling uncovers a p53-associated role for microRNA-31 in inhibiting the proliferation of serous ovarian carcinomas and other cancers. Cancer Res. 2010; 70:1906–1915. PMID: 20179198.
Article17. Wei J, Wang Z, Wang Z, Yang Y, Fu C, Zhu J, et al. MicroRNA-31 function as a suppressor was regulated by epigenetic mechanisms in gastric cancer. Biomed Res Int. 2017; 2017:5348490. PMID: 29333444.
Article18. Valastyan S, Weinberg RA. miR-31: a crucial overseer of tumor metastasis and other emerging roles. Cell Cycle. 2010; 9:2124–2129. PMID: 20505365.
Article19. Jiao F, Hu H, Han T, Zhuo M, Yuan C, Yang H, et al. Aberrant expression of nuclear HDAC3 and cytoplasmic CDH1 predict a poor prognosis for patients with pancreatic cancer. Oncotarget. 2016; 7:16505–16516. PMID: 26918727.
Article20. Koumangoye RB, Andl T, Taubenslag KJ, Zilberman ST, Taylor CJ, Loomans HA, et al. SOX4 interacts with EZH2 and HDAC3 to suppress microRNA-31 in invasive esophageal cancer cells. Mol Cancer. 2015; 14:24. PMID: 25644061.
Article21. Bayat S, Shekari Khaniani M, Choupani J, Alivand MR, Mansoori Derakhshan S. HDACis (class I), cancer stem cell, and phytochemicals: cancer therapy and prevention implications. Biomed Pharmacother. 2018; 97:1445–1453. PMID: 29156535.
Article22. Zhang M, Yin Y, Dorfman RG, Zou T, Pan Y, Li Y, et al. Down-regulation of HDAC3 inhibits growth of cholangiocarcinoma by inducing apoptosis. Oncotarget. 2017; 8:99402–99413. PMID: 29245911.
Article23. Marks P, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. Histone deacetylases and cancer: causes and therapies. Nat Rev Cancer. 2001; 1:194–202. PMID: 11902574.
Article24. Paszkowski J, Whitham SA. Gene silencing and DNA methylation processes. Curr Opin Plant Biol. 2001; 4:123–129. PMID: 11228434.
Article25. Liu Z, Bai J, Zhang L, Lou F, Ke F, Cai W, et al. Conditional knockout of microRNA-31 promotes the development of colitis associated cancer. Biochem Biophys Res Commun. 2017; 490:62–68. PMID: 28600172.
Article26. Stepicheva NA, Song JL. Function and regulation of microRNA-31 in development and disease. Mol Reprod Dev. 2016; 83:654–674. PMID: 27405090.
Article27. Rasheed SA, Teo CR, Beillard EJ, Voorhoeve PM, Zhou W, Ghosh S, et al. MicroRNA-31 controls G protein alpha-13 (GNA13) expression and cell invasion in breast cancer cells. Mol Cancer. 2015; 14:67. PMID: 25889182.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- c-Myc-Induced Long Non-Coding RNA Small Nucleolar RNA Host Gene 7 Regulates Glycolysis in Breast Cancer
- HDAC1 Expression in Invasive Ductal Carcinoma of the Breast and Its Value as a Good Prognostic Factor
- Anti-Cancer Effect of 3-(4-dimethylamino phenyl)-N-hydroxy-2-propenamide in MCF-7 Human Breast Cancer
- The Role of Histone Acetylation in Mesenchymal Stem Cell Differentiation
- Epigenetic Modifications: Novel Therapeutic Approach for Thyroid Cancer