Yonsei Med J.
2018 May;59(3):431-437. 10.3349/ymj.2018.59.3.431.
Phenotypic Analysis of Korean Patients with Abnormal Chromosomal Microarray in Patients with Unexplained Developmental Delay/Intellectual Disability
- Affiliations
-
- 1Department of Pediatrics, Gachon University Gil Medical Center, Incheon, Korea.
- 2Department of Rehabilitation Medicine, Konyang University College of Medicine, Daejeon, Korea.
- 3Department of Pediatrics, Konyang University College of Medicine, Daejeon, Korea.
- 4Green Cross Genome, Yongin, Korea.
- 5Department of Medical Genetics, Konyang University College of Medicine, Daejeon, Korea. raredisease@hanmail.net
- KMID: 2407867
- DOI: http://doi.org/10.3349/ymj.2018.59.3.431
Abstract
- PURPOSE
The present study aimed to investigate chromosomal microarray (CMA) and clinical data in patients with unexplained developmental delay/intellectual disability (DD/ID) accompanying dysmorphism, congenital anomalies, or epilepsy. We also aimed to evaluate phenotypic clues in patients with pathogenic copy number variants (CNVs).
MATERIALS AND METHODS
We collected clinical and CMA data from patients at Konyang University Hospital between September 2013 and October 2014. We included patients who had taken the CMA test to evaluate the etiology of unexplained DD/ID.
RESULTS
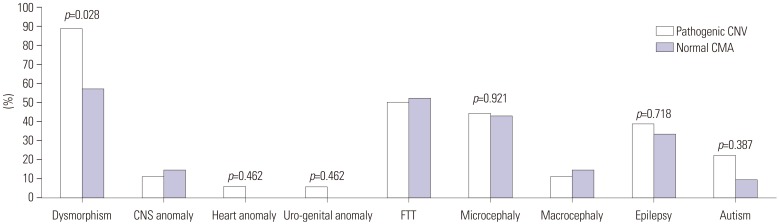
All of the 50 patients identified had DD/ID. Thirty-nine patients had dysmorphism, 19 patients suffered from epilepsy, and 12 patients had congenital anomalies. Twenty-nine of the 50 patients (58%) showed abnormal results. Eighteen (36%) were considered to have pathogenic CNVs. Dysmorphism (p=0.028) was significantly higher in patients with pathogenic CNVs than in those with normal CMA. Two or more clinical features were presented by 61.9% (13/21) of the patients with normal CMA and by 83.3% (15/18) of the patients with pathogenic CMA.
CONCLUSION
Dysmorphism can be a phenotypic clue to pathogenic CNVs. Furthermore, pathogenic CNV might be more frequently found if patients have two or more clinical features in addition to DD/ID.
MeSH Terms
Figure
Reference
-
1. Leonard H, Wen X. The epidemiology of mental retardation: challenges and opportunities in the new millennium. Ment Retard Dev Disabil Res Rev. 2002; 8:117–134. PMID: 12216056.
Article2. Vissers LE, Gilissen C, Veltman JA. Genetic studies in intellectual disability and related disorders. Nat Rev Genet. 2016; 17:9–18. PMID: 26503795.
Article3. Rauch A, Hoyer J, Guth S, Zweier C, Kraus C, Becker C, et al. Diagnostic yield of various genetic approaches in patients with unexplained developmental delay or mental retardation. Am J Med Genet A. 2006; 140:2063–2074. PMID: 16917849.
Article4. Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010; 86:749–764. PMID: 20466091.
Article5. Battaglia A, Doccini V, Bernardini L, Novelli A, Loddo S, Capalbo A, et al. Confirmation of chromosomal microarray as a first-tier clinical diagnostic test for individuals with developmental delay, intellectual disability, autism spectrum disorders and dysmorphic features. Eur J Paediatr Neurol. 2013; 17:589–599. PMID: 23711909.
Article6. Zilina O, Teek R, Tammur P, Kuuse K, Yakoreva M, Vaidla E, et al. Chromosomal microarray analysis as a first-tier clinical diagnostic test: Estonian experience. Mol Genet Genomic Med. 2014; 2:166–175. PMID: 24689080.7. Chong WW, Lo IF, Lam ST, Wang CC, Luk HM, Leung TY, et al. Performance of chromosomal microarray for patients with intellectual disabilities/developmental delay, autism, and multiple congenital anomalies in a Chinese cohort. Mol Cytogenet. 2014; 7:34. PMID: 24926319.
Article8. Howell KB, Kornberg AJ, Harvey AS, Ryan MM, Mackay MT, Freeman JL, et al. High resolution chromosomal microarray in undiagnosed neurological disorders. J Paediatr Child Health. 2013; 49:716–724. PMID: 23731025.
Article9. Byeon JH, Shin E, Kim GH, Lee K, Hong YS, Lee JW, et al. Application of array-based comparative genomic hybridization to pediatric neurologic diseases. Yonsei Med J. 2014; 55:30–36. PMID: 24339284.
Article10. Iourov IY, Vorsanova SG, Kurinnaia OS, Zelenova MA, Silvanovich AP, Yurov YB. Molecular karyotyping by array CGH in a Russian cohort of children with intellectual disability, autism, epilepsy and congenital anomalies. Mol Cytogenet. 2012; 5:46. PMID: 23272938.
Article11. Uwineza A, Caberg JH, Hitayezu J, Hellin AC, Jamar M, Dideberg V, et al. Array-CGH analysis in Rwandan patients presenting development delay/intellectual disability with multiple congenital anomalies. BMC Med Genet. 2014; 15:79. PMID: 25016475.
Article12. Coulter ME, Miller DT, Harris DJ, Hawley P, Picker J, Roberts AE, et al. Chromosomal microarray testing influences medical management. Genet Med. 2011; 13:770–776. PMID: 21716121.
Article13. Palmer E, Speirs H, Taylor PJ, Mullan G, Turner G, Einfeld S, et al. Changing interpretation of chromosomal microarray over time in a community cohort with intellectual disability. Am J Med Genet A. 2014; 164A:377–385. PMID: 24311194.
Article14. Kim HJ, Cho E, Park JB, Im WY, Kim HJ. A Korean family with KBG syndrome identified by ANKRD11 mutation, and phenotypic comparison of ANKRD11 mutation and 16q24.3 microdeletion. Eur J Med Genet. 2015; 58:86–94. PMID: 25464108.
Article15. Gilissen C, Hehir-Kwa JY, Thung DT, van de Vorst M, van Bon BW, Willemsen MH, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014; 511:344–347. PMID: 24896178.
Article16. Girirajan S, Brkanac Z, Coe BP, Baker C, Vives L, Vu TH, et al. Relative burden of large CNVs on a range of neurodevelopmental phenotypes. PLoS Genet. 2011; 7:e1002334. PMID: 22102821.
Article17. Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011; 43:838–846. PMID: 21841781.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Chromosomal Microarray Testing in 42 Korean Patients with Unexplained Developmental Delay, Intellectual Disability, Autism Spectrum Disorders, and Multiple Congenital Anomalies
- Chromosomal Microarray With Clinical Diagnostic Utility in Children With Developmental Delay or Intellectual Disability
- The first Korean case of 2p15p16.1 microdeletion syndrome, characterized by facial dysmorphism, developmental delay, and congenital hypothyroidism
- Diagnostic distal 16p11.2 deletion in a preterm infant with facial dysmorphism
- The First Cases of OPHN1 Exons 1 and 2 Deletion in Two X-linked Intellectual Developmental Disorder Patients in Korea