Ann Lab Med.
2018 May;38(3):242-248. 10.3343/alm.2018.38.3.242.
Spectrum of MNX1 Pathogenic Variants and Associated Clinical Features in Korean Patients with Currarino Syndrome
- Affiliations
-
- 1Department of Laboratory Medicine, Kangbuk Samsung Hospital, Sungkyunkwan University School of Medicine, Seoul, Korea.
- 2Department of Laboratory Medicine, Seoul National University College of Medicine, Seoul, Korea.
- 3Department of Laboratory Medicine, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea.
- 4Department of Laboratory Medicine, Gyeongsang National University Changwon Hospital, Changwon, Korea.
- 5Department of Laboratory Medicine, Seoul National University Bundang Hospital, Seongnam, Korea.
- 6Department of Pediatric Surgery, Seoul National University College of Medicine, Seoul, Korea. spkhy02@snu.ac.kr
- KMID: 2403458
- DOI: http://doi.org/10.3343/alm.2018.38.3.242
Abstract
- BACKGROUND
The major genetic cause of Currarino syndrome (CS), a congenital malformation syndrome typically characterized by sacral agenesis, anorectal malformation, and presence of a pre-sacral mass, is known to be pathogenic variants in motor neuron and pancreas homeobox 1 (MNX1), which exist in almost all familial cases and 30% of sporadic cases. Less commonly, a large deletion or a complex rearrangement involving the 7q36 region is associated with CS. We investigated the spectrum of MNX1 pathogenic variants and associated clinical features in the Korean patients with CS.
METHODS
We enrolled 25 patients with CS, including 24 sporadic cases and one familial case. Direct sequencing of MNX1 and multiplex ligation-dependent probe amplification were performed. We also analyzed clinical phenotypes and evaluated genotype-phenotype correlations.
RESULTS
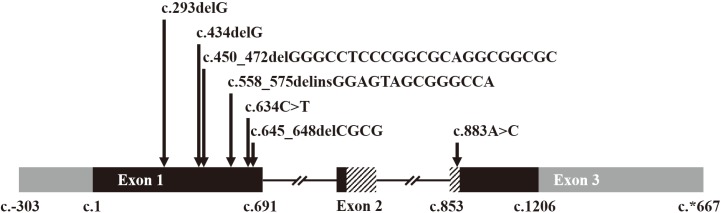
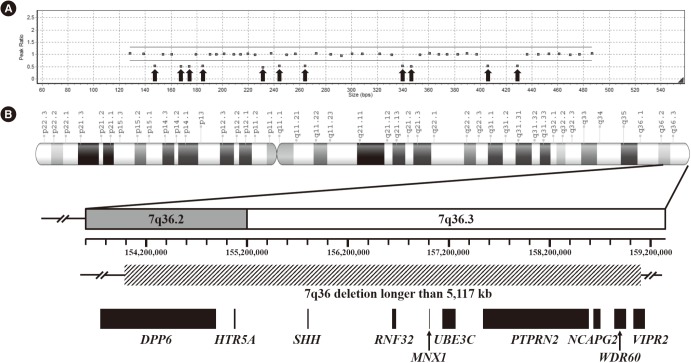
We identified six novel variants amongst a total of six null variants, one missense variant, and one large deletion. The null variants included four frameshift variants (p.Gly98Alafs*124, p.Gly145Alafs*77, p.Gly151Leufs*67, and p.Ala216Profs*5) and two nonsense variants (p.Tyr186* and p.Gln212*). The missense variant, p.Lys295Gln, was located in the highly-conserved homeobox domain and was predicted to be deleterious. A large deletion involving the 7q36 region was detected in one patient. Pathogenic variants in MNX1 were detected in 28% of all CS cases and 25% of sporadic cases. The clinical phenotype was variable in patients with and without pathogenic variants; no significant genotype-phenotype correlation was observed.
CONCLUSIONS
This study revealed the spectrum and phenotypic variability of MNX1 pathogenic variants in the Korean population.
Keyword
MeSH Terms
Figure
Reference
-
1. Currarino G, Coln D, Votteler T. Triad of anorectal, sacral, and presacral anomalies. AJR Am J Roentgenol. 1981; 137:395–398. PMID: 6789651.2. Horn D, Tonnies H, Neitzel H, Wahl D, Hinkel GK, von Moers A, et al. Minimal clinical expression of the holoprosencephaly spectrum and of Currarino syndrome due to different cytogenetic rearrangements deleting the Sonic Hedgehog gene and the HLXB9 gene at 7q36.3. Am J Med Genet A. 2004; 128A:85–92. PMID: 15211664.3. Pavone P, Ruggieri M, Lombardo I, Sudi J, Biancheri R, Castellano-Chiodo D, et al. Microcephaly, sensorineural deafness and Currarino triad with duplication-deletion of distal 7q. Eur J Pediatr. 2010; 169:475–481. PMID: 19838731.4. Cuturilo G, Hodge JC, Runke CK, Thorland EC, Al-Owain MA, Ellison JW, et al. Phenotype analysis impacts testing strategy in patients with Currarino syndrome. Clin Genet. 2016; 89:109–114. PMID: 25691298.5. Hagan DM, Ross AJ, Strachan T, Lynch SA, Ruiz-Perez V, Wang YM, et al. Mutation analysis and embryonic expression of the HLXB9 Currarino syndrome gene. Am J Hum Genet. 2000; 66:1504–1515. PMID: 10749657.6. Kochling J, Karbasiyan M, Reis A. Spectrum of mutations and genotype-phenotype analysis in Currarino syndrome. Eur J Hum Genet. 2001; 9:599–605. PMID: 11528505.7. Wang RY, Jones JR, Chen S, Rogers RC, Friez MJ, Schwartz CE, et al. A previously unreported mutation in a Currarino syndrome kindred. Am J Med Genet A. 2006; 140:1923–1930. PMID: 16906559.8. Cretolle C, Sarnacki S, Amiel J, Genevieve D, Encha-Razavi F, Zrelli S, et al. Currarino syndrome shown by prenatal onset ventriculomegaly and spinal dysraphism. Am J Med Genet A. 2007; 143A:871–874. PMID: 17352395.9. Kim IS, Oh SY, Choi SJ, Kim JH, Park KH, Park HK, et al. Clinical and genetic analysis of HLXB9 gene in Korean patients with Currarino syndrome. J Hum Genet. 2007; 52:698–701. PMID: 17612791.10. Zu S, Winberg J, Arnberg F, Palmer G, Svensson PJ, Wester T, et al. Mutation analysis of the motor neuron and pancreas homeobox 1 (MNX1, former HLXB9) gene in Swedish patients with Currarino syndrome. J Pediatr Surg. 2011; 46:1390–1395. PMID: 21763840.11. Markljung E, Adamovic T, Cao J, Naji H, Kaiser S, Wester T, et al. Novel mutations in the MNX1 gene in two families with Currarino syndrome and variable phenotype. Gene. 2012; 507:50–53. PMID: 22820079.12. Wang Y, Wu Y. A novel HLXB9 mutation in a Chinese family with Currarino syndrome. Eur J Pediatr Surg. 2012; 22:243–245. PMID: 21960426.13. Yates VD, Wilroy RS, Whitington GL, Simmons JC. Anterior sacral defects: an autosomal dominantly inherited condition. J Pediatr. 1983; 102:239–242. PMID: 6822928.14. Lynch SA, Bond PM, Copp AJ, Kirwan WO, Nour S, Balling R, et al. A gene for autosomal dominant sacral agenesis maps to the holoprosencephaly region at 7q36. Nat Genet. 1995; 11:93–95. PMID: 7550324.15. Ross AJ, Ruiz-Perez V, Wang Y, Hagan DM, Scherer S, Lynch SA, et al. A homeobox gene, HLXB9, is the major locus for dominantly inherited sacral agenesis. Nat Genet. 1998; 20:358–361. PMID: 9843207.16. Belloni E, Martucciello G, Verderio D, Ponti E, Seri M, Jasonni V, et al. Involvement of the HLXB9 homeobox gene in Currarino syndrome. Am J Hum Genet. 2000; 66:312–319. PMID: 10631160.17. Cretolle C, Pelet A, Sanlaville D, Zerah M, Amiel J, Jaubert F, et al. Spectrum of HLXB9 gene mutations in Currarino syndrome and genotype-phenotype correlation. Hum Mutat. 2008; 29:903–910. PMID: 18449898.18. Merello E, De Marco P, Ravegnani M, Riccipetitoni G, Cama A, Capra V. Novel MNX1 mutations and clinical analysis of familial and sporadic Currarino cases. Eur J Med Genet. 2013; 56:648–654. PMID: 24095820.19. Kim AY, Yoo SY, Kim JH, Eo H, Jeon TY. Currarino syndrome: variable imaging features in three siblings with HLXB9 gene mutation. Clin Imaging. 2013; 37:398–402. PMID: 23466002.20. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010; 7:248–249. PMID: 20354512.21. Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009; 4:1073–1081. PMID: 19561590.22. Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014; 11:361–362. PMID: 24681721.23. Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009; 37:e67. PMID: 19339519.24. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015; 17:405–424. PMID: 25741868.25. Garcia-Barcelo M, So MT, Lau DK, Leon TY, Yuan ZW, Cai WS, et al. Population differences in the polyalanine domain and 6 new mutations in HLXB9 in patients with Currarino syndrome. Clin Chem. 2006; 52:46–52. PMID: 16254195.26. Garcia-Barcelo MM, Lui VC, So MT, Miao X, Leon TY, Yuan ZW, et al. MNX1 (HLXB9) mutations in Currarino patients. J Pediatr Surg. 2009; 44:1892–1898. PMID: 19853743.27. Benzacken B, Siffroi JP, Le Bourhis C, Krabchi K, Joye N, Maschino F, et al. Different proximal and distal rearrangements of chromosome 7q associated with holoprosencephaly. J Med Genet. 1997; 34:899–903. PMID: 9391882.28. Coutton C, Poreau B, Devillard F, Durand C, Odent S, Rozel C, et al. Currarino syndrome and HPE microform associated with a 2.7-Mb deletion in 7q36.3 excluding SHH gene. Mol Syndromol. 2014; 5:25–31. PMID: 24550762.29. Frints SG, Schoenmakers EF, Smeets E, Petit P, Fryns JP. De novo 7q36 deletion: breakpoint analysis and types of holoprosencephaly. Am J Med Genet. 1998; 75:153–158. PMID: 9450876.30. Frints SG, Schrander-Stumpel CT, Schoenmakers EF, Engelen JJ, Reekers AB, van den Neucker AM, et al. Strong variable clinical presentation in 3 patients with 7q terminal deletion. Genet Couns. 1998; 9:5–14. PMID: 9555580.31. Holm I, Monclair T, Lundar T, Stadheim B, Prescott TE, Eiklid KL. A 5.8 kb deletion removing the entire MNX1 gene in a Norwegian family with Currarino syndrome. Gene. 2013; 518:457–460. PMID: 23370340.32. Masuno M, Fukushima Y, Sugio Y, Ikeda M, Kuroki Y. Two unrelated cases of single maxillary central incisor with 7q terminal deletion. Jinrui Idengaku Zasshi. 1990; 35:311–317. PMID: 2094780.33. Rodriguez L, Cuadrado Perez I, Herrera Montes J, Lorente Jareno ML, Lopez Grondona F, Martinez-Frias ML. Terminal deletion of the chromosome 7(q36-qter) in an infant with sacral agenesis and anterior myelomeningocele. Am J Med Genet. 2002; 110:73–77. PMID: 12116275.34. Roessler E, Ward DE, Gaudenz K, Belloni E, Scherer SW, Donnai D, et al. Cytogenetic rearrangements involving the loss of the Sonic Hedgehog gene at 7q36 cause holoprosencephaly. Hum Genet. 1997; 100:172–181. PMID: 9254845.35. Lee SK. Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron. 2003; 38:731–745. PMID: 12797958.36. Lee SK, Jurata LW, Funahashi J, Ruiz EC, Pfaff SL. Analysis of embryonic motoneuron gene regulation: derepression of general activators function in concert with enhancer factors. Development. 2004; 131:3295–3306. PMID: 15201216.37. Nakano T, Windrem M, Zappavigna V, Goldman SA. Identification of a conserved 125 base-pair Hb9 enhancer that specifies gene expression to spinal motor neurons. Dev Biol. 2005; 283:474–485. PMID: 15913596.38. Holm I, Spildrejorde M, Stadheim B, Eiklid KL, Samarakoon PS. Whole exome sequencing of sporadic patients with Currarino Syndrome: a report of three trios. Gene. 2017; 624:50–55. PMID: 28456592.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Currarino triad with Mullerian duct anomaly in mother and daughter without MNX1 gene mutation
- Clinical Characteristics and Treatment of Currarino Syndrome: A Single Institutional Experience
- Germline Variants in MLH1, MSH2, and MSH6 in Korean Patients with Lynch Syndrome
- Presacral Mass in Currarino Triad: Case Report
- A Case of Currarino Triad