Ann Lab Med.
2018 May;38(3):196-203. 10.3343/alm.2018.38.3.196.
Clinical Utility of a Diagnostic Approach to Detect Genetic Abnormalities in Multiple Myeloma: A Single Institution Experience
- Affiliations
-
- 1Division of Hematology-Oncology, Department of Medicine, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea. kihyunkimk@gmail.com
- 2Department of Internal Medicine, Hallym University Medical Center, Hallym University College of Medicine, Dongtan, Korea.
- 3Department of Laboratory Medicine and Genetics, Soonchunhyang University Bucheon Hospital, Soonchunhyang University College of Medicine, Bucheon, Korea.
- 4Department of Laboratory Medicine & Genetics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea. drsunnyhk@gmail.com
- KMID: 2403452
- DOI: http://doi.org/10.3343/alm.2018.38.3.196
Abstract
- BACKGROUND
The identification of genetic abnormalities in patients with multiple myeloma (MM) has gained emphasis because genetics-based risk stratification significantly affects overall survival (OS). We investigated genetic abnormalities using conventional cytogenetics and FISH and analyzed the prognostic significance of the identified additional abnormalities in MM.
METHODS
In total, 267 bone marrow samples were collected from February 2006 to November 2013 from patients who were newly diagnosed as having MM in a tertiary-care hospital in Korea. The clinical and laboratory data were retrospectively obtained. Cox proportional hazard regression was used to examine the relationship between clinical/genetic factors and survival outcome, using univariate and multivariate models.
RESULTS
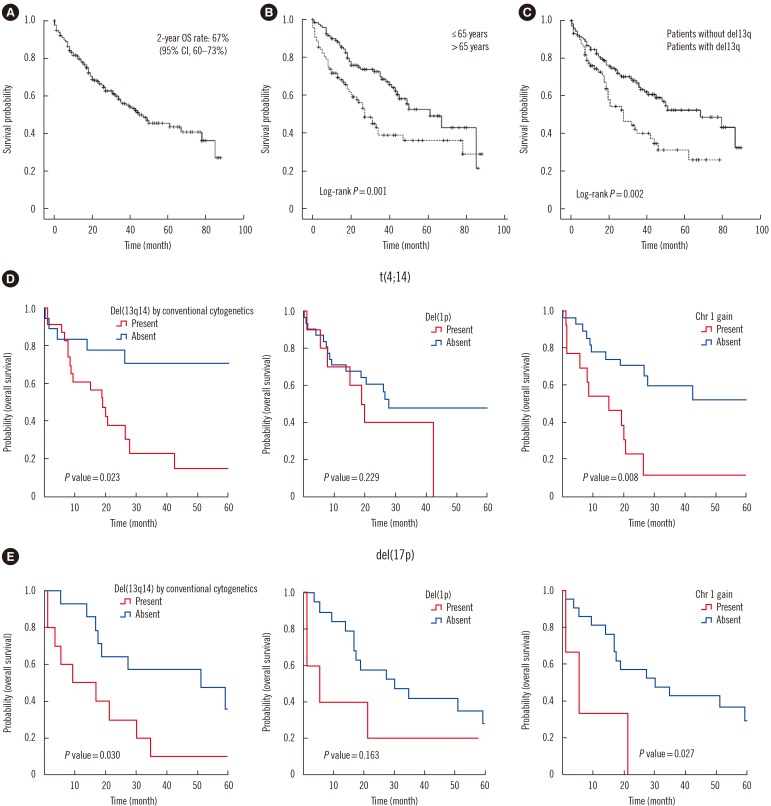
Using conventional cytogenetic analysis and FISH, 45% (120/267) and 69% (183/267) patients, respectively, were identified to harbor genetic abnormalities. In the univariate analysis, the following genetic variables were identified to affect OS: abnormal karyotype (P < 0.001), aneuploidy (P=0.046), −13 or del(13q) (P=0.002), 1q amplification (P < 0.001), and t(4;14) (P=0.020). In the multivariate analysis, the presence of −13 or del(13q) was the only significant genetic factor affecting OS (P=0.012) with a hazard ratio (HR) of 2.131 (95% confidence interval [CI], 1.185-3.832) in addition to the clinical factor of age (>65 years) (P=0.013) with an HR of 2.505 (95% CI, 1.218-5.151).
CONCLUSIONS
Our findings highlight the importance of applying a comprehensive approach for detecting genetic abnormalities, which could be closely associated with the prognostic significance of MM.
MeSH Terms
Figure
Cited by 1 articles
-
Clinical Utility of Next-Generation Flow-Based Minimal Residual Disease Assessment in Patients with Multiple Myeloma
Hyun-Young Kim, In Young Yoo, Dae Jin Lim, Hee-Jin Kim, Sun-Hee Kim, Sang Eun Yoon, Seok Jin Kim, Duck Cho, Kihyun Kim
Ann Lab Med. 2022;42(5):558-565. doi: 10.3343/alm.2022.42.5.558.
Reference
-
1. Swerdlow SH, Campo E, editors. WHO classification of tumours of haematopoietic and lymphoid tissues. 4th ed. Lyon: International Agency for Research on Cancer;2008. p. 200–207.2. Chng WJ, Dispenzieri A, Chim CS, Fonseca R, Goldschmidt H, Lentzsch S, et al. IMWG consensus on risk stratification in multiple myeloma. Leukemia. 2014; 28:269–277. PMID: 23974982.3. Fonseca R, Bergsagel PL, Drach J, Shaughnessy J, Gutierrez N, Stewart AK, et al. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009; 23:2210–2221. PMID: 19798094.4. Munshi NC, Anderson KC, Bergsagel PL, Shaughnessy J, Palumbo A, Durie B, et al. Consensus recommendations for risk stratification in multiple myeloma: report of the International Myeloma Workshop Consensus Panel 2. Blood. 2011; 117:4696–4700. PMID: 21292777.5. Bergsagel PL, Kuehl WM. Chromosome translocations in multiple myeloma. Oncogene. 2001; 20:5611–5622. PMID: 11607813.6. Avet-Loiseau H, Attal M, Moreau P, Charbonnel C, Garban F, Hulin C, et al. Genetic abnormalities and survival in multiple myeloma: the experience of the Intergroupe Francophone du Myelome. Blood. 2007; 109:3489–3495. PMID: 17209057.7. Bergsagel PL, Kuehl WM. Molecular pathogenesis and a consequent classification of multiple myeloma. J Clin Oncol. 2005; 23:6333–6338. PMID: 16155016.8. Calasanz MJ, Cigudosa JC, Odero MD, Ferreira C, Ardanaz MT, Fraile A, et al. Cytogenetic analysis of 280 patients with multiple myeloma and related disorders: primary breakpoints and clinical correlations. Genes Chromosomes Cancer. 1997; 18:84–93. PMID: 9115968.9. Dong H, Yang HS, Jagannath S, Stephenson CF, Brenholz P, Mazumder A, et al. Risk stratification of plasma cell neoplasm: insights from plasma cell-specific cytoplasmic immunoglobulin fluorescence in situ hybridization (cIg FISH) vs. conventional FISH. Clin Lymphoma Myeloma Leuk. 2012; 12:366–374. PMID: 22658896.10. Gersen SL, Keagle MB, editors. The Principles of Clinical Cytogenetics. 2nd ed. Totowa, NJ: Humana Press;2004. p. 64–65.11. Wolff DJ, Bagg A, Cooley LD, Dewald GW, Hirsch BA, Jacky PB, et al. Guidance for fluorescence in situ hybridization testing in hematologic disorders. J Mol Diagn. 2007; 9:134–143. PMID: 17384204.12. Lee JW, Lee JK, Hong YJ, Hong SI, Chang YH. Correlation of chromosomal aberrations with prognostic markers in multiple myeloma patients--a single institution study. Korean J Lab Med. 2008; 28:413–418. PMID: 19127104.13. Jekarl DW, Min CK, Kwon A, Kim H, Chae H, Kim M, et al. Impact of genetic abnormalities on the prognoses and clinical parameters of patients with multiple myeloma. Ann Lab Med. 2013; 33:248–254. PMID: 23826560.14. Bang SM, Kim YR, Cho HI, Chi HS, Seo EJ, Park CJ, et al. Identification of 13q deletion, trisomy 1q, and IgH rearrangement as the most frequent chromosomal changes found in Korean patients with multiple myeloma. Cancer Genet Cytogenet. 2006; 168:124–132. PMID: 16843102.15. Lim JH, Seo EJ, Park CJ, Jang S, Chi HS, Suh C, et al. Cytogenetic classification in Korean multiple myeloma patients: prognostic significance of hyperdiploidy with 47-50 chromosomes and the number of structural abnormalities. Eur J Haematol. 2014; 92:313–320. PMID: 24372944.16. Jacobus SJ, Kumar S, Uno H, van Wier SA, Ahmann GJ, Henderson KJ, et al. Impact of high-risk classification by FISH: an eastern cooperative oncology group (ECOG) study E4A03. Br J Haematol. 2011; 155:340–348. PMID: 21902684.17. Durie BG, Salmon SE. A clinical staging system for multiple myeloma. Correlation of measured myeloma cell mass with presenting clinical features, response to treatment, and survival. Cancer. 1975; 36:842–854. PMID: 1182674.18. Choi JH, Yoon JH, Yang SK. Clinical value of new staging systems for multiple myeloma. Cancer Res Treat. 2007; 39:171–174. PMID: 19746184.19. Avet-Loiseau H, Durie BG, Cavo M, Attal M, Gutierrez N, Haessler J, et al. Combining fluorescent in situ hybridization data with ISS staging improves risk assessment in myeloma: an International Myeloma Working Group collaborative project. Leukemia. 2013; 27:711–717. PMID: 23032723.20. Oh S, Koo DH, Kwon MJ, Kim K, Suh C, Min CK, et al. Chromosome 13 deletion and hypodiploidy on conventional cytogenetics are robust prognostic factors in Korean multiple myeloma patients: web-based multicenter registry study. Ann Hematol. 2014; 93:1353–1361. PMID: 24671365.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Correlation of Chromosomal Aberrations with Prognostic Markers in Multiple Myeloma Patients- A Single Institution Study
- Multiple Myeloma and Epidural Spinal Cord Compression : Case Presentation and a Spine Surgeon's Perspective
- Liver Involvement of Multiple Myeloma Mimicking Intrahepatic Cholangiocarcinoma: A Case Report
- A retrospective analysis of cytogenetic alterations in patients with newly diagnosed multiple myeloma: a single center study in Korea
- Multiple myeloma