J Pathol Transl Med.
2018 Jan;52(1):37-44. 10.4132/jptm.2017.10.20.
The Smad4/PTEN Expression Pattern Predicts Clinical Outcomes in Colorectal Adenocarcinoma
- Affiliations
-
- 1Department of Pathology, Hanyang University College of Medicine, Seoul, Korea. kyueng@gmail.com sspaik@hanyang.ac.kr
- 2Department of Pathology, Kangbuk Samsung Hospital, Sungkyunkwan University School of Medicine, Seoul, Korea.
- KMID: 2403256
- DOI: http://doi.org/10.4132/jptm.2017.10.20
Abstract
- BACKGROUND
Smad4 and PTEN are prognostic indicators for various tumor types. Smad4 regulates tumor suppression, whereas PTEN inhibits cell proliferation. We analyzed and compared the performance of Smad4 and PTEN for predicting the prognosis of patients with colorectal adenocarcinoma.
METHODS
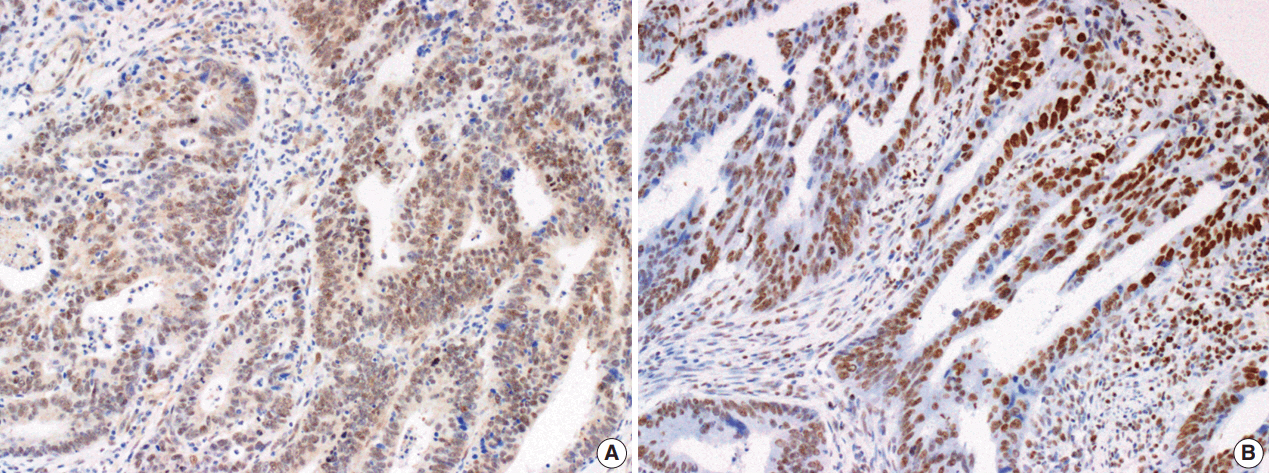
Combined expression patterns based on Smad4+/- and PTEN+/- status were evaluated by immunostaining using a tissue microarray of colorectal adenocarcinoma. The relationships between the protein expression and clinicopathological variables were analyzed.
RESULTS
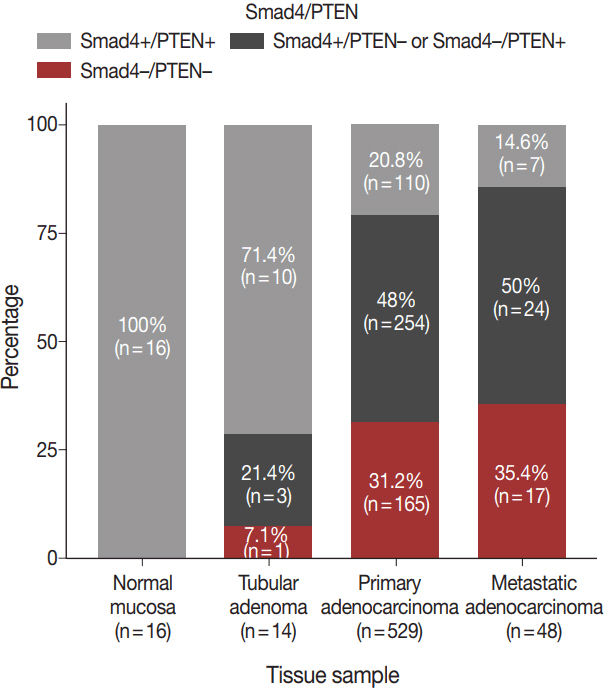
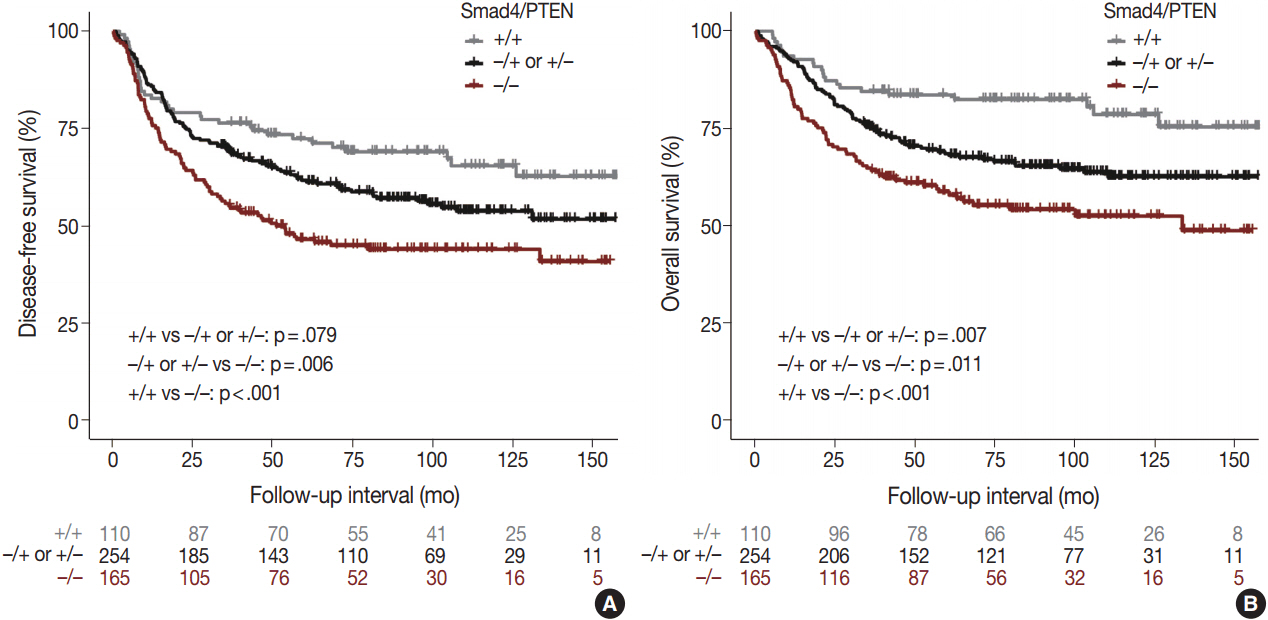
Smad4-/PTEN- status was most frequently observed in metastatic adenocarcinoma, followed by primary adenocarcinoma and tubular adenoma (p<.001). When Smad4-/PTEN- and Smad4+/PTEN+ groups were compared, Smad4-/PTEN- status was associated with high N stage (p=.018) and defective mismatch repair proteins (p=.006). Significant differences in diseasefree survival and overall survival were observed among the three groups (Smad4+/PTEN+, Smad4-/PTEN+ or Smad4+/PTEN-, and Smad4-/PTEN-) (all p<.05).
CONCLUSIONS
Concurrent loss of Smad4 and PTEN may lead to more aggressive disease and poor prognosis in patients with colorectal adenocarcinoma compared to the loss of Smad4 or PTEN alone.
Keyword
MeSH Terms
Figure
Cited by 1 articles
-
Clinicopathological Characterization and Prognostic Implication of SMAD4 Expression in Colorectal Carcinoma
Seung-Yeon Yoo, Ji-Ae Lee, Yunjoo Shin, Nam-Yun Cho, Jeong Mo Bae, Gyeong Hoon Kang
J Pathol Transl Med. 2019;53(5):289-297. doi: 10.4132/jptm.2019.06.07.
Reference
-
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015; 65:87–108.
Article2. Cunningham D, Atkin W, Lenz HJ, et al. Colorectal cancer. Lancet. 2010; 375:1030–47.
Article3. Vogelstein B, Fearon ER, Hamilton SR, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988; 319:525–32.
Article4. Valcz G, Sipos F, Krenács T, et al. Increase of alpha-SMA(+) and CK (+) cells as an early sign of epithelial-mesenchymal transition during colorectal carcinogenesis. Pathol Oncol Res. 2012; 18:371–6.5. Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993; 363:558–61.
Article6. Peltomäki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J Clin Oncol. 2003; 21:1174–9.
Article7. Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010; 138:87.e3.
Article8. Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007; 318:1108–13.9. Starr TK, Allaei R, Silverstein KA, et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009; 323:1747–50.
Article10. Segditsas S, Tomlinson I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene. 2006; 25:7531–7.
Article11. Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015; 21:1350–6.12. Choi D, Lee HW, Hur KY, et al. Cancer stem cell markers CD133 and CD24 correlate with invasiveness and differentiation in colorectal adenocarcinoma. World J Gastroenterol. 2009; 15:2258–64.
Article13. Jing F, Kim HJ, Kim CH, Kim YJ, Lee JH, Kim HR. Colon cancer stem cell markers CD44 and CD133 in patients with colorectal cancer and synchronous hepatic metastases. Int J Oncol. 2015; 46:1582–8.
Article14. Sahlberg SH, Spiegelberg D, Glimelius B, Stenerlöw B, Nestor M. Evaluation of cancer stem cell markers CD133, CD44, CD24: association with AKT isoforms and radiation resistance in colon cancer cells. PLoS One. 2014; 9:e94621.
Article15. Chen KL, Pan F, Jiang H, et al. Highly enriched CD133(+)CD44(+) stem-like cells with CD133(+)CD44(high) metastatic subset in HCT116 colon cancer cells. Clin Exp Metastasis. 2011; 28:751–63.
Article16. Kechagioglou P, Papi RM, Provatopoulou X, et al. Tumor suppressor PTEN in breast cancer: heterozygosity, mutations and protein expression. Anticancer Res. 2014; 34:1387–400.17. Saal LH, Gruvberger-Saal SK, Persson C, et al. Recurrent gross mutations of the PTEN tumor suppressor gene in breast cancers with deficient DSB repair. Nat Genet. 2008; 40:102–7.
Article18. Wang X, Huang H, Young KH. The PTEN tumor suppressor gene and its role in lymphoma pathogenesis. Aging (Albany NY). 2015; 7:1032–49.
Article19. Zhang LL, Mu GG, Ding QS, et al. Phosphatase and tensin homolog (PTEN) represses colon cancer progression through inhibiting paxillin transcription via PI3K/AKT/NF-kappaB pathway. J Biol Chem. 2015; 290:15018–29.20. Park SH, Won J, Kim SI, et al. Molecular testing of brain tumor. J Pathol Transl Med. 2017; 51:205–23.
Article21. Miyaki M, Kuroki T. Role of Smad4 (DPC4) inactivation in human cancer. Biochem Biophys Res Commun. 2003; 306:799–804.
Article22. Xu X, Kobayashi S, Qiao W, et al. Induction of intrahepatic cholangiocellular carcinoma by liver-specific disruption of Smad4 and Pten in mice. J Clin Invest. 2006; 116:1843–52.23. Chow JY, Cabral JA, Chang J, Carethers JM. TGFbeta modulates PTEN expression independently of SMAD signaling for growth proliferation in colon cancer cells. Cancer Biol Ther. 2008; 7:1694–9.24. Ding Z, Wu CJ, Chu GC, et al. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011; 470:269–73.25. Teng Y, Sun AN, Pan XC, et al. Synergistic function of Smad4 and PTEN in suppressing forestomach squamous cell carcinoma in the mouse. Cancer Res. 2006; 66:6972–81.
Article26. Yan P, Klingbiel D, Saridaki Z, et al. Reduced expression of SMAD4 is associated with poor survival in colon cancer. Clin Cancer Res. 2016; 22:3037–47.
Article27. Liu J, Cho SN, Akkanti B, et al. ErbB2 pathway activation upon Smad4 loss promotes lung tumor growth and metastasis. Cell Rep. 2015; 10:1599–613.
Article28. Xu X, Ehdaie B, Ohara N, Yoshino T, Deng CX. Synergistic action of Smad4 and PTEN in suppressing pancreatic ductal adenocarcinoma formation in mice. Oncogene. 2010; 29:674–86.
Article29. Jang KS, Song YS, Jang SH, et al. Clinicopathological significance of nuclear PTEN expression in colorectal adenocarcinoma. Histopathology. 2010; 56:229–39.
Article30. Peppercorn J, Perou CM, Carey LA. Molecular subtypes in breast cancer evaluation and management: divide and conquer. Cancer Invest. 2008; 26:1–10.
Article31. Stambolic V, Suzuki A, de la Pompa JL, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998; 95:29–39.
Article32. Davies MA, Koul D, Dhesi H, et al. Regulation of Akt/PKB activity, cellular growth, and apoptosis in prostate carcinoma cells by MMAC/PTEN. Cancer Res. 1999; 59:2551–6.33. Haeger SM, Thompson JJ, Kalra S, et al. Smad4 loss promotes lung cancer formation but increases sensitivity to DNA topoisomerase inhibitors. Oncogene. 2016; 35:577–86.
Article34. Ahmed S, Bradshaw AD, Gera S, Dewan MZ, Xu R. The TGF-beta/Smad4 signaling pathway in pancreatic carcinogenesis and its clinical significance. J Clin Med. 2017; 6:5.35. Tachibana M, Shibakita M, Ohno S, et al. Expression and prognostic significance of PTEN product protein in patients with esophageal squamous cell carcinoma. Cancer. 2002; 94:1955–60.
Article36. Lee HS, Lee HK, Kim HS, Yang HK, Kim WH. Tumour suppressor gene expression correlates with gastric cancer prognosis. J Pathol. 2003; 200:39–46.
Article37. Li J, Yen C, Liaw D, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997; 275:1943–7.38. Zhao Y, Zheng R, Li J, Lin F, Liu L. Loss of phosphatase and tensin homolog expression correlates with clinicopathological features of non-small cell lung cancer patients and its impact on survival: a systematic review and meta-analysis. Thorac Cancer. 2017; 8:203–13.
Article39. Risinger JI, Hayes AK, Berchuck A, Barrett JC. PTEN/MMAC1 mutations in endometrial cancers. Cancer Res. 1997; 57:4736–8.40. Yang L, Kuang LG, Zheng HC, et al. PTEN encoding product: a marker for tumorigenesis and progression of gastric carcinoma. World J Gastroenterol. 2003; 9:35–9.
Article41. Dicuonzo G, Angeletti S, Garcia-Foncillas J, et al. Colorectal carcinomas and PTEN/MMAC1 gene mutations. Clin Cancer Res. 2001; 7:4049–53.42. Guanti G, Resta N, Simone C, et al. Involvement of PTEN mutations in the genetic pathways of colorectal cancerogenesis. Hum Mol Genet. 2000; 9:283–7.
Article43. Reynisdóttir I, Polyak K, Iavarone A, Massagué J. Kip/Cip and Ink4 Cdk inhibitors cooperate to induce cell cycle arrest in response to TGF-beta. Genes Dev. 1995; 9:1831–45.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Smad4 Expression in Gastric Adenocarcinoma
- Clinicopathological Significance of SMAD4 Expression in Breast Cancer
- Loss of PTEN Expression in Primary Lung Cancer
- Clinical Significance of PTEN Expression in Colorectal Cancer
- Clinicopathological Characterization and Prognostic Implication of SMAD4 Expression in Colorectal Carcinoma