J Vet Sci.
2018 Jan;19(1):99-106. 10.4142/jvs.2018.19.1.99.
Integrated analysis of microRNA and mRNA expressions in peripheral blood leukocytes of Warmblood horses before and after exercise
- Affiliations
-
- 1Laboratory of Veterinary Clinical Pathology, College of Veterinary Medicine, Seoul National University, Seoul 08826, Korea. yongbaek@snu.ac.kr
- 2Laboratory of Environmental Health and Biomarkers, College of Veterinary Medicine, Seoul National University, Seoul 08826, Korea.
- 3BK21 PLUS Program for Creative Veterinary Science Research, College of Veterinary Medicine, Seoul National University, Seoul 08826, Korea.
- 4Research Institute for Veterinary Science, College of Veterinary Medicine, Seoul National University, Seoul 08826, Korea.
- KMID: 2402700
- DOI: http://doi.org/10.4142/jvs.2018.19.1.99
Abstract
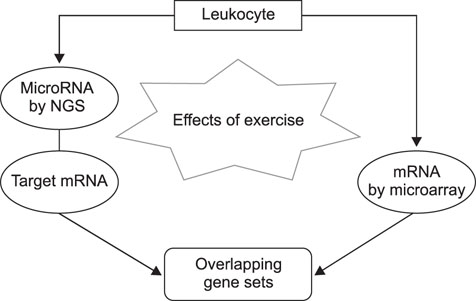
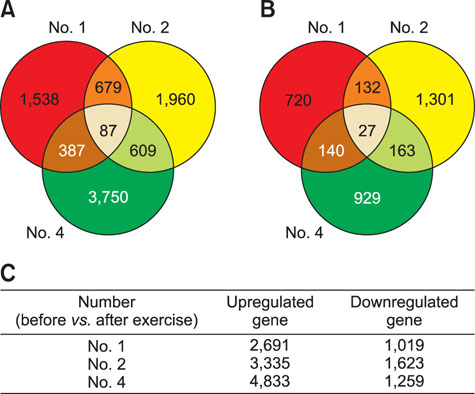
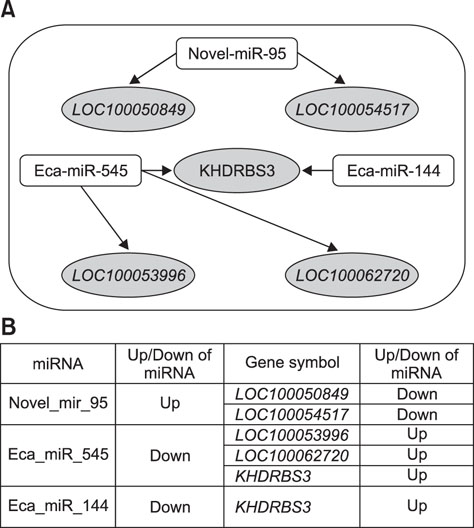
- Exercise capacity is a valuable trait in horses, and it has been used as a horse selection criterion. Although exercise affects molecular homeostasis and adaptation in horses, the mechanisms underlying these effects are not fully described. This study was carried out to identify changes in the blood profiles of microRNAs (miRNAs) and mRNAs induced by exercise in horse leukocytes. Total RNAs isolated from the peripheral blood leukocytes of four Warmblood horses before and after exercise were subjected to next-generation sequencing (NGS) and microarray analyses to determine the miRNA and mRNA expression profiles, respectively. The expressions of 6 miRNAs, including 4 known and 2 novel miRNAs, were altered by exercise. The predicted target genes of the differentially expressed miRNAs identified by NGS were matched to the exercise-induced mRNAs determined by microarray analysis. Five genes (LOC100050849, LOC100054517, KHDRBS3, LOC100053996, and LOC100062720) from the microarray analysis were matched to the predicted target genes of the 6 miRNAs. The subset of mRNAs and miRNAs affected by exercise in peripheral blood leukocytes may be useful in elucidating the molecular mechanisms of exercise-associated physiology in horses.
Keyword
MeSH Terms
Figure
Reference
-
1. Aschenbach WG, Ho RC, Sakamoto K, Fujii N, Li Y, Kim YB, Hirshman MF, Goodyear LJ. Regulation of dishevelled and beta-catenin in rat skeletal muscle: an alternative exercise-induced GSK-3beta signaling pathway. Am J Physiol Endocrinol Metab. 2006; 291:E152–E158.2. Barrey E. Genetics and genomics in equine exercise physiology: an overview of the new applications of molecular biology as positive and negative markers of performance and health. Equine Vet J Suppl. 2010; (38):561–568.3. Barrey E, Mucher E, Robert C, Amiot F, Gidrol X. Gene expression profiling in blood cells of endurance horses completing competition or disqualified due to metabolic disorder. Equine Vet J Suppl. 2006; (36):43–49.
Article4. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995; 57:289–300.
Article5. Calderone A, Murphy RJ, Lavoie J, Colombo F, Béliveau L. TGF-beta(1) and prepro-ANP mRNAs are differentially regulated in exercise-induced cardiac hypertrophy. J Appl Physiol (1985). 2001; 91:771–776.
Article6. Clarkson PM, Devaney JM, Gordish-Dressman H, Thompson PD, Hubal MJ, Urso M, Price TB, Angelopoulos TJ, Gordon PM, Moyna NM, Pescatello LS, Visich PS, Zoeller RF, Seip RL, Hoffman EP. ACTN3 genotype is associated with increases in muscle strength in response to resistance training in women. J Appl Physiol (1985). 2005; 99:154–163.
Article7. Dey S, Singh RH, Dey PK. Exercise training: significance of regional alterations in serotonin metabolism of rat brain in relation to antidepressant effect of exercise. Physiol Behav. 1992; 52:1095–1099.
Article8. Dillies MA, Rau A, Aubert J, Hennequet-Antier C, Jeanmougin M, Servant N, Keime C, Marot G, Castel D, Estelle J, Guernec G, Jagla B, Jouneau L, Laloë D, Le Gall C, Schaëffer B, Le Crom S, Guedj M, Jaffrézic F. French StatOmique Consortium. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief Bioinform. 2013; 14:671–683.
Article9. Ding Q, Vaynman S, Akhavan M, Ying Z, Gomez-Pinilla F. Insulin-like growth factor I interfaces with brain-derived neurotrophic factor-mediated synaptic plasticity to modulate aspects of exercise-induced cognitive function. Neuroscience. 2006; 140:823–833.
Article10. Fink L, Hölschermann H, Kwapiszewska G, Muyal JP, Lengemann B, Bohle RM, Santoso S. Characterization of platelet-specific mRNA by real-time PCR after laser-assisted microdissection. Thromb Haemost. 2003; 90:749–756.
Article11. Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008; 9:831–842.
Article12. Gaffney B, Cunningham EP. Estimation of genetic trend in racing performance of thoroughbred horses. Nature. 1988; 332:722–724.
Article13. Gim JA, Ayarpadikannan S, Eo J, Kwon YJ, Choi Y, Lee HK, Park KD, Yang YM, Cho BW, Kim HS. Transcriptional expression changes of glucose metabolism genes after exercise in thoroughbred horses. Gene. 2014; 547:152–158.
Article14. Gu J, Orr N, Park SD, Katz LM, Sulimova G, MacHugh DE, Hill EW. A genome scan for positive selection in thoroughbred horses. PLoS One. 2009; 4:e5767.
Article15. Heinemeier K, Langberg H, Olesen JL, Kjaer M. Role of TGF-beta1 in relation to exercise-induced type I collagen synthesis in human tendinous tissue. J Appl Physiol (1985). 2003; 95:2390–2397.
Article16. Hill EW, Katz LM, MacHugh DE. Genomics of performance. In : Chowdhary BP, editor. Equine Genomics. Oxford: Blackwell Publishing;2013. p. 265–283.17. Kim MC, Lee SW, Ryu DY, Cui FJ, Bhak J, Kim Y. Identification and characterization of microRNAs in normal equine tissues by next generation sequencing. PLoS One. 2014; 9:e93662.
Article18. Kulikova MA, Maliuchenko NV, Timofeeva MA, Shleptsova VA, Tschegol'kova IuA, Vediakov AM, Tonevitskiĭ AG. [Polymorphisms of the main genes of neurotransmitter systems: I. the dopaminergic system]. Fiziol Cheloveka. 2007; 33:105–112.
Article19. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25.
Article20. Lee S, Hwang S, Yu HJ, Oh D, Choi YJ, Kim MC, Kim Y, Ryu DY. Expression of microRNAs in horse plasma and their characteristic nucleotide composition. PLoS One. 2016; 11:e0146374.
Article21. Levine MA. Investigating the origins of horse domestication. Equine Vet J Suppl. 1999; (28):6–14.
Article22. Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008; 24:713–714.
Article23. Ma Z, Qi J, Meng S, Wen B, Zhang J. Swimming exercise training-induced left ventricular hypertrophy involves microRNAs and synergistic regulation of the PI3K/AKT/mTOR signaling pathway. Eur J Appl Physiol. 2013; 113:2473–2486.
Article24. Maxwell PH, Dachs GU, Gleadle JM, Nicholls LG, Harris AL, Stratford IJ, Hankinson O, Pugh CW, Ratcliffe PJ. Hypoxia-inducible factor-1 modulates gene expression in solid tumors and influences both angiogenesis and tumor growth. Proc Natl Acad Sci U S A. 1997; 94:8104–8109.
Article25. McCue ME, Valberg SJ, Lucio M, Mickelson JR. Glycogen synthase 1 (GYS1) mutation in diverse breeds with polysaccharide storage myopathy. J Vet Intern Med. 2008; 22:1228–1233.
Article26. McGivney BA, Eivers SS, MacHugh DE, MacLeod JN, O'Gorman GM, Park SD, Katz LM, Hill EW. Transcriptional adaptations following exercise in thoroughbred horse skeletal muscle highlights molecular mechanisms that lead to muscle hypertrophy. BMC Genomics. 2009; 10:638.
Article27. Mounier R, Pialoux V, Cayre A, Schmitt L, Richalet JP, Robach P, Lasne F, Roels B, Millet G, Coudert J, Clottes E, Fellmann N. Leukocyte's Hif-1 expression and training-induced erythropoietic response in swimmers. Med Sci Sports Exerc. 2006; 38:1410–1417.
Article28. Mounier R, Pialoux V, Roels B, Thomas C, Millet G, Mercier J, Coudert J, Fellmann N, Clottes E. Effect of intermittent hypoxic training on HIF gene expression in human skeletal muscle and leukocytes. Eur J Appl Physiol. 2009; 105:515–524.
Article29. Murphy BA, Wagner AL, McGlynn OF, Kharazyan F, Browne JA, Elliott JA. Exercise influences circadian gene expression in equine skeletal muscle. Vet J. 2014; 201:39–45.
Article30. Norman B, Sabina RL, Jansson E. Regulation of skeletal muscle ATP catabolism by AMPD1 genotype during sprint exercise in asymptomatic subjects. J Appl Physiol (1985). 2001; 91:258–264.
Article31. Park KD, Park J, Ko J, Kim BC, Kim HS, Ahn K, Do KT, Choi H, Kim HM, Song S, Lee S, Jho S, Kong HS, Yang YM, Jhun BH, Kim C, Kim TH, Hwang S, Bhak J, Lee HK, Cho BW. Whole transcriptome analyses of six thoroughbred horses before and after exercise using RNA-Seq. BMC Genomics. 2012; 13:473.
Article32. Radom-Aizik S, Zaldivar F, Haddad F, Cooper DM. Impact of brief exercise on peripheral blood NK cell gene and microRNA expression in young adults. J Appl Physiol (1985). 2013; 114:628–636.
Article33. Radom-Aizik S, Zaldivar F Jr, Leu SY, Adams GR, Oliver S, Cooper DM. Effects of exercise on microRNA expression in young males peripheral blood mononuclear cells. Clin Transl Sci. 2012; 5:32–38.
Article34. Russell AP. Molecular regulation of skeletal muscle mass. Clin Exp Pharmacol Physiol. 2010; 37:378–384.
Article35. Schröder W, Klostermann A, Distl O. Candidate genes for physical performance in the horse. Vet J. 2011; 190:39–48.
Article36. Tonevitsky AG, Maltseva DV, Abbasi A, Samatov TR, Sakharov DA, Shkurnikov MU, Lebedev AE, Galatenko VV, Grigoriev AI, Northoff H. Dynamically regulated miRNA-mRNA networks revealed by exercise. BMC Physiol. 2013; 13:9.
Article37. Walmsley SR, Print C, Farahi N, Peyssonnaux C, Johnson RS, Cramer T, Sobolewski A, Condliffe AM, Cowburn AS, Johnson N, Chilvers ER. Hypoxia-induced neutrophil survival is mediated by HIF-1alpha-dependent NF-kappaB activity. J Exp Med. 2005; 201:105–115.
Article38. Yeh SH, Chuang H, Lin LW, Hsiao CY, Eng HL. Regular tai chi chuan exercise enhances functional mobility and CD4CD25 regulatory T cells. Br J Sports Med. 2006; 40:239–243.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A novel biomarker of exercise-induced stress in horses
- Peripheral serotoninergic response to physical exercise in athletic horses
- Integrated Analysis of MicroRNA and mRNA Expression Profiles in Rheumatoid Arthritis Synovial Monocytes
- Speckle-tracking analysis of myocardial deformation in correlation to age in healthy horses
- Comparison of Interleukin-8 Levels in Long-Distance Runners and Healthy Sedentary Non-Athletic Control Subjects