Intest Res.
2018 Jan;16(1):90-98. 10.5217/ir.2018.16.1.90.
Distribution of cytomegalovirus genotypes among ulcerative colitis patients in Okinawa, Japan
- Affiliations
-
- 1Department of Infectious, Respiratory, and Digestive Medicine, Graduate School of Medicine, University of the Ryukyus, Okinawa, Japan.
- 2Department of Endoscopy, Graduate School of Medicine, University of the Ryukyus, Okinawa, Japan. hokama-a@med.u-ryukyu.ac.jp
- KMID: 2402652
- DOI: http://doi.org/10.5217/ir.2018.16.1.90
Abstract
- BACKGROUND/AIMS
To determine the prevalence of glycoprotein B (gB), glycoprotein N (gN), and glycoprotein H (gH) genotypes of human cytomegalovirus (HCMV) superimposed on ulcerative colitis (UC) patients in Japan.
METHODS
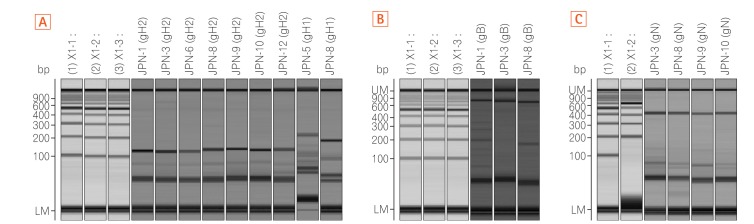
Four archived stool samples and 7-archived extracted DNA from stool samples of 11 UC patients with positive multiplex polymerase chain reaction (PCR) results for HCMV were used UL55 gene encoding gB, UL73 gene encoding gN, and UL75 gene encoding gH were identified by PCR. Genotypes of gB and glycoprotein N were determined by sequencing.
RESULTS
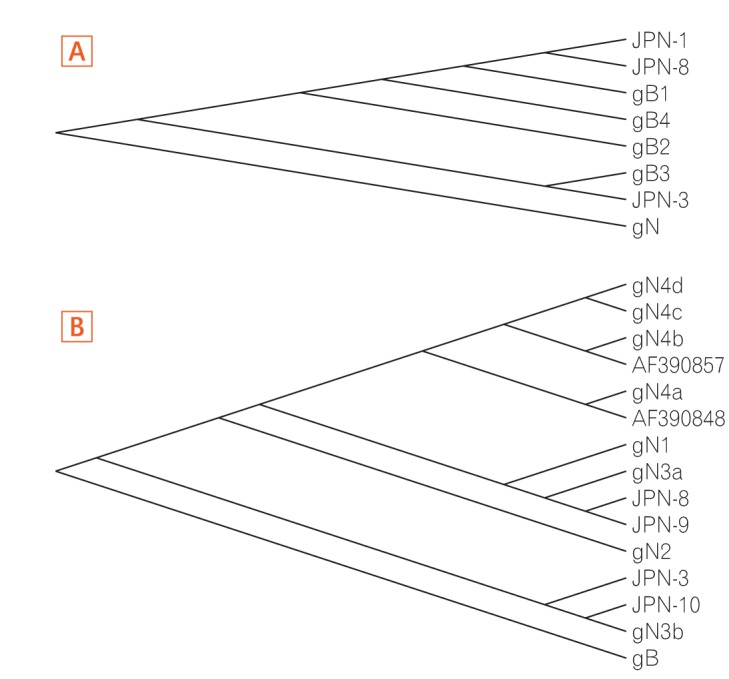
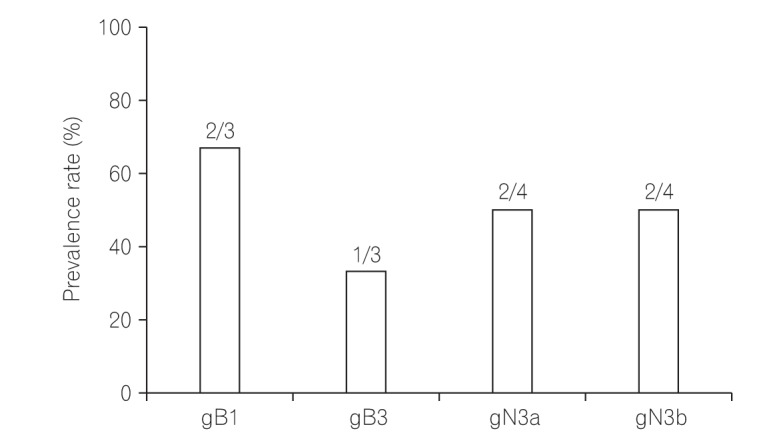
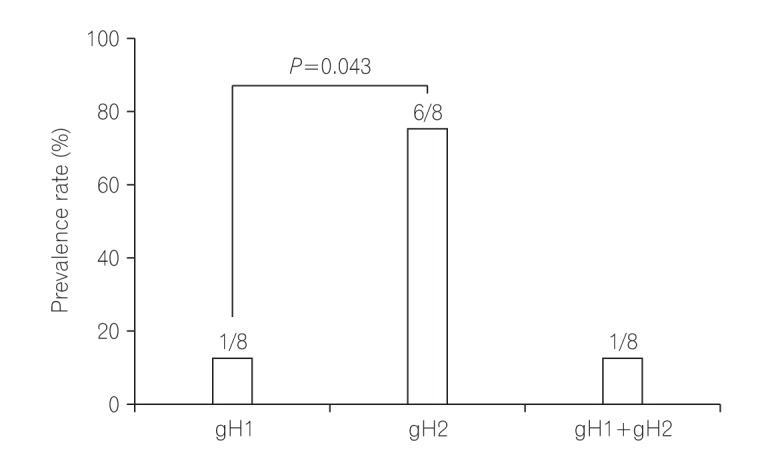
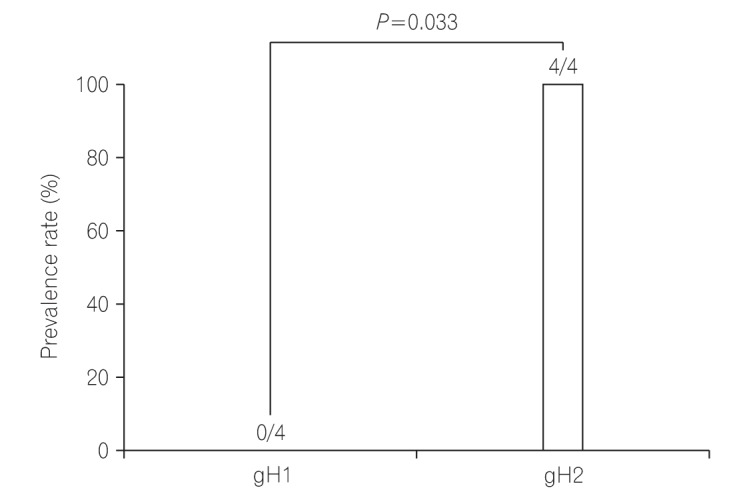
Among 11 samples, 8 samples were amplified through PCR. gB, gN, and gH genotypes were successfully detected in 3 of 8 (37.5%), 4 of 8 (50%), and 8 of 8 (100%), respectively. The distribution of gB and gN genotypes analyzed through phylogenetic analysis were as follows: gB1 (2/3, 66.7%), gB3 (1/3, 33.3%), gN3a (2/4, 50%), and gN3b (2/4, 50%). Other gB genotypes (gB2 and gB4) and gN genotypes (gN1, gN2, and gN4) were not detected in this study. Out of successfully amplified 8 samples of gH genotype, gH1 and gH2 were distributed in 12.5% and 75% samples, respectively. Only 1 sample revealed mixed infection of gH genotype. The distribution of gH1 and gH2 differed significantly (1:6, P < 0.05) in UC patients. The distribution of single gH genotype also revealed significant difference in UC patients who were treated with immunosuppressive drug (P < 0.05).
CONCLUSIONS
In this study, gB1, gN3, and gH2 gene were determined as the most frequently observed genotypes in UC patients, which suggest that there might be an association between these genotypes of HCMV and UC.
MeSH Terms
Figure
Reference
-
1. Murphy E, Shenk T. Human cytomegalovirus genome. Curr Top Microbiol Immunol. 2008; 325:1–19. PMID: 18637497.
Article2. Chou SW, Dennison KM. Analysis of interstrain variation in cytomegalovirus glycoprotein B sequences encoding neutralization-related epitopes. J Infect Dis. 1991; 163:1229–1234. PMID: 1709960.
Article3. Burkhardt C, Himmelein S, Britt W, Winkler T, Mach M. Glycoprotein N subtypes of human cytomegalovirus induce a strain-specific antibody response during natural infection. J Gen Virol. 2009; 90(Pt 8):1951–1961. PMID: 19420160.
Article4. Paterson DA, Dyer AP, Milne RS, Sevilla-Reyes E, Gompels UA. A role for human cytomegalovirus glycoprotein O (gO) in cell fusion and a new hypervariable locus. Virology. 2002; 293:281–294. PMID: 11886248.
Article5. Vanarsdall AL, Ryckman BJ, Chase MC, Johnson DC. Human cytomegalovirus glycoproteins gB and gH/gL mediate epithelial cell-cell fusion when expressed either in cis or in trans. J Virol. 2008; 82:11837–11850. PMID: 18815310.
Article6. Rasmussen L, Geissler A, Cowan C, Chase A, Winters M. The genes encoding the gCIII complex of human cytomegalovirus exist in highly diverse combinations in clinical isolates. J Virol. 2002; 76:10841–10848. PMID: 12368327.
Article7. Meyer-König U, Haberland M, von Laer D, Haller O, Hufert FT. Intragenic variability of human cytomegalovirus glycoprotein B in clinical strains. J Infect Dis. 1998; 177:1162–1169. PMID: 9592998.
Article8. Pignatelli S, Dal Monte P, Landini MP. gpUL73 (gN) genomic variants of human cytomegalovirus isolates are clustered into four distinct genotypes. J Gen Virol. 2001; 82(Pt 11):2777–2784. PMID: 11602789.
Article9. Chou S. Molecular epidemiology of envelope glycoprotein H of human cytomegalovirus. J Infect Dis. 1992; 166:604–607. PMID: 1323623.
Article10. Görzer I, Kerschner H, Redlberger-Fritz M, Puchhammer-Stöckl E. Human cytomegalovirus (HCMV) genotype populations in immunocompetent individuals during primary HCMV infection. J Clin Virol. 2010; 48:100–103. PMID: 20362496.
Article11. Mujtaba G, Khurshid A, Sharif S, et al. Distribution of cytomegalovirus genotypes among neonates born to infected mothers in Islamabad, Pakistan. PLoS One. 2016; 11:e0156049. DOI: 10.1371/journal.pone.0156049. PMID: 27367049.
Article12. Dieamant DC, Bonon SH, Peres RM, et al. Cytomegalovirus (CMV) genotype in allogeneic hematopoietic stem cell transplantation. BMC Infect Dis. 2013; 13:310. PMID: 23841715.
Article13. Nakase H, Matsumura K, Yoshino T, Chiba T. Systematic review: cytomegalovirus infection in inflammatory bowel disease. J Gastroenterol. 2008; 43:735–740. PMID: 18958541.
Article14. Lawlor G, Moss AC. Cytomegalovirus in inflammatory bowel disease: pathogen or innocent bystander? Inflamm Bowel Dis. 2010; 16:1620–1627. PMID: 20232408.
Article15. Nahar S, Iraha A, Hokama A, et al. Evaluation of a multiplex PCR assay for detection of cytomegalovirus in stool samples from patients with ulcerative colitis. World J Gastroenterol. 2015; 21:12667–12675. PMID: 26640344.
Article16. Schroeder KW, Tremaine WJ, Ilstrup DM. Coated oral 5-aminosalicylic acid therapy for mildly to moderately active ulcerative colitis: a randomized study. N Engl J Med. 1987; 317:1625–1629. PMID: 3317057.
Article17. Paca-Uccaralertkun S, Hiatt R, Leecharoen R, Tan-Ariya P, Mungthin M, Pongphong S. Human cytomegalovirus gB1 genotypes among children who live at the Phayathai Babies' home in Nonthaburi, Thailand. Southeast Asian J Trop Med Public Health. 2013; 44:636–640. PMID: 24050097.18. Vilibic-Cavlek T, Kolaric B, Ljubin-Sternak S, Kos M, Kaic B, Mlinaric-Galinovic G. Prevalence and dynamics of cytomegalovirus infection among patients undergoing chronic hemodialysis. Indian J Nephrol. 2015; 25:95–98. PMID: 25838647.
Article19. Santos CA, Brennan DC, Fraser VJ, Olsen MA. Incidence, risk factors, and outcomes of delayed-onset cytomegalovirus disease in a large, retrospective cohort of heart transplant recipients. Transplant Proc. 2014; 46:3585–3592. PMID: 25498094.
Article20. Heiden D, Tun N, Maningding E, et al. Training clinicians treating HIV to diagnose cytomegalovirus retinitis. Bull World Health Organ. 2014; 92:903–908. PMID: 25552774.
Article21. Cottone M, Pietrosi G, Martorana G, et al. Prevalence of cytomegalovirus infection in severe refractory ulcerative and Crohn's colitis. Am J Gastroenterol. 2001; 96:773–775. PMID: 11280549.
Article22. Matsuoka K, Iwao Y, Mori T, et al. Cytomegalovirus is frequently reactivated and disappears without antiviral agents in ulcerative colitis patients. Am J Gastroenterol. 2007; 102:331–337. PMID: 17156136.
Article23. Kandiel A, Lashner B. Cytomegalovirus colitis complicating inflammatory bowel disease. Am J Gastroenterol. 2006; 101:2857–2865. PMID: 17026558.
Article24. Pignatelli S, Dal Monte P, Rossini G, Landini MP. Genetic polymorphisms among human cytomegalovirus (HCMV) wild-type strains. Rev Med Virol. 2004; 14:383–410. PMID: 15386592.
Article25. Ikuta K, Minematsu T, Inoue N, et al. Cytomegalovirus (CMV) glycoprotein H-based serological analysis in Japanese healthy pregnant women, and in neonates with congenital CMV infection and their mothers. J Clin Virol. 2013; 58:474–478. PMID: 23916379.
Article26. Vogel JU, Otte J, Koch F, Gümbel H, Doerr HW, Cinatl J Jr. Role of human cytomegalovirus genotype polymorphisms in AIDS patients with cytomegalovirus retinitis. Med Microbiol Immunol. 2013; 202:37–47. PMID: 22669631.
Article27. Madi N, Al-Nakib W, Pacsa A, Saeed T. Cytomegalovirus genotypes gB1 and gH1 are the most predominant genotypes among renal transplant recipients in Kuwait. Transplant Proc. 2011; 43:1634–1637. PMID: 21693248.
Article28. Fries BC, Chou S, Boeckh M, Torok-Storb B. Frequency distribu-tion of cytomegalovirus envelope glycoprotein genotypes in bone marrow transplant recipients. J Infect Dis. 1994; 169:769–774. PMID: 8133090.
Article29. Fan J, Zhang X, Chen XM, et al. Monitoring of human cytomegalovirus glycoprotein B genotypes using real-time quantitative PCR in immunocompromised Chinese patients. J Virol Methods. 2009; 160:74–77. PMID: 19406161.
Article30. Zhang MG, Wang HB, Wang YZ, Pan Q. Human cytomegalovirus glycoprotein B genotypes in congenitally infected neonates. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2011; 25:262–264. PMID: 22097601.31. Taherkhani R, Farshadpour F, Makvandi M, et al. Determination of cytomegalovirus prevalence and glycoprotein B genotypes among ulcerative colitis patients in ahvaz, iran. Jundishapur J Microbiol. 2015; 8:e17458. DOI: 10.5812/jjm.17458. PMID: 25793098.
Article32. Nogueira E, Ozaki KS, Tomiyama H, Câmara NO, Granato CF. Clinical correlations of human cytomegalovirus strains and viral load in kidney transplant recipients. Int Immunopharmacol. 2009; 9:26–31. PMID: 18824137.
Article33. Chen JY, Zheng TL, Zhou T, et al. Human cytomegalovirus prevalence and distribution of glycoprotein B, O genotypes among hospitalized children with respiratory infections in West China, 2009-2014. Trop Med Int Health. 2016; 21:1428–1434. PMID: 27515771.
Article34. Xia CS, Zhang Z. Analysis of human cytomegalovirus glycoprotein N genotypes in Chinese hematopoietic stem cell transplant recipients. Arch Virol. 2011; 156:17–23. PMID: 20878191.
Article35. Mohamed HT, El-Shinawi M, Nouh MA, et al. Inflammatory breast cancer: high incidence of detection of mixed human cytomegalovirus genotypes associated with disease pathogenesis. Front Oncol. 2014; 4:246. PMID: 25309872.
Article36. Rossini G, Pignatelli S, Dal Monte P, et al. Monitoring for human cytomegalovirus infection in solid organ transplant recipients through antigenemia and glycoprotein N (gN) variants: evidence of correlation and potential prognostic value of gN genotypes. Microbes Infect. 2005; 7:890–896. PMID: 15878684.
Article37. Pignatelli S, Rossini G, Dal Monte P, Gatto MR, Landini MP. Human cytomegalovirus glycoprotein N genotypes in AIDS patients. AIDS. 2003; 17:761–763. PMID: 12646803.
Article38. Puchhammer-Stöckl E, Görzer I, Zoufaly A, et al. Emergence of multiple cytomegalovirus strains in blood and lung of lung transplant recipients. Transplantation. 2006; 81:187–194. PMID: 16436961.
Article39. Pignatelli S, Lazzarotto T, Gatto MR, et al. Cytomegalovirus gN genotypes distribution among congenitally infected newborns and their relationship with symptoms at birth and sequelae. Clin Infect Dis. 2010; 51:33–41. PMID: 20504230.
Article40. Pignatelli S, Dal Monte P, Rossini G, et al. Human cytomegalovirus glycoprotein N (gpUL73-gN) genomic variants: identification of a novel subgroup, geographical distribution and evidence of positive selective pressure. J Gen Virol. 2003; 84(Pt 3):647–655. PMID: 12604817.
Article41. de Vries JJ, Wessels E, Korver AM, et al. Rapid genotyping of cytomegalovirus in dried blood spots by multiplex real-time PCR assays targeting the envelope glycoprotein gB and gH genes. J Clin Microbiol. 2012; 50:232–237. PMID: 22116158.
Article42. Guo S, Yu MM, Li G, Zhou H, Fang F, Shu SN. Studies on genotype of human cytomegalovirus glycoprotein H from infantile clinical isolates. Zhonghua Er Ke Za Zhi. 2013; 51:260–264. PMID: 23927798.43. Yan H, Koyano S, Inami Y, et al. Genetic linkage among human cytomegalovirus glycoprotein N (gN) and gO genes, with evidence for recombination from congenitally and post-natally infected Japanese infants. J Gen Virol. 2008; 89(Pt 9):2275–2279. PMID: 18753237.
Article44. Paradowska E, Jabłońska A, Studzińska M, et al. Cytomegalovirus glycoprotein H genotype distribution and the relationship with hearing loss in children. J Med Virol. 2014; 86:1421–1427. PMID: 24615599.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A Case of Cytomegalvirus Colitis Developed during the Treatment of Ulcerative Colitis
- A Clinical Significance of Assessing Cytomegalovirus Infection Status in Patients With Ulcerative Colitis
- Cytomegalovirus Colitis with Ulcerative Colitis in the Steroid Naive Immunocompetent Patient
- The Cumulative Colectomy Rate in Patients with Cytomegalovirus-Positive Ulcerative Colitis
- Fecal Microbiota Transplantation to Patients with Refractory Very Early Onset Ulcerative Colitis