Intest Res.
2017 Oct;15(4):495-501. 10.5217/ir.2017.15.4.495.
Is methylation analysis of SFRP2, TFPI2, NDRG4, and BMP3 promoters suitable for colorectal cancer screening in the Korean population?
- Affiliations
-
- 1Division of Gastroenterology, Department of Internal Medicine, Kangbuk Samsung Hospital, Sungkyunkwan University School of Medicine, Seoul, Korea. diksmc.park@samsung.com
- 2Gastrointestinal Cancer Center, Kangbuk Samsung Hospital, Sungkyunkwan University School of Medicine, Seoul, Korea.
- 3Comprehensive Health Care Center, Korea Cancer Center Hospital, Korea Institute of Radiological & Medical Sciences, Seoul, Korea.
- 4Department of Surgery, Kangbuk Samsung Hospital, Sungkyunkwan University School of Medicine, Seoul, Korea.
- 5Department of Pathology, Kangbuk Samsung Hospital, Sungkyunkwan University School of Medicine, Seoul, Korea.
- KMID: 2396399
- DOI: http://doi.org/10.5217/ir.2017.15.4.495
Abstract
- BACKGROUND/AIMS
Colorectal cancer (CRC) screening using stool DNA was recently found to yield good detection rates. A multi-target stool DNA test (Cologuard®, Exact Sciences), including methylated genes has been recently approved by the U.S. Food and Drug Administration. The aim of this study was to validate these aberrantly methylated genes as stool-based DNA markers for detecting CRC and colorectal advanced adenoma (AA) in the Korean population.
METHODS
A single-center study was conducted in 36 patients with AA; 35 patients with CRC; and 40 endoscopically diagnosed healthy controls using CRC screening colonoscopy. The methylation status of the SFRP2, TFPI2, NDRG4, and BMP3 promoters was investigated blindly using bisulfate-modified stool DNA obtained from 111 participants. Methylation status was investigated by methylation-specific polymerase chain reaction.
RESULTS
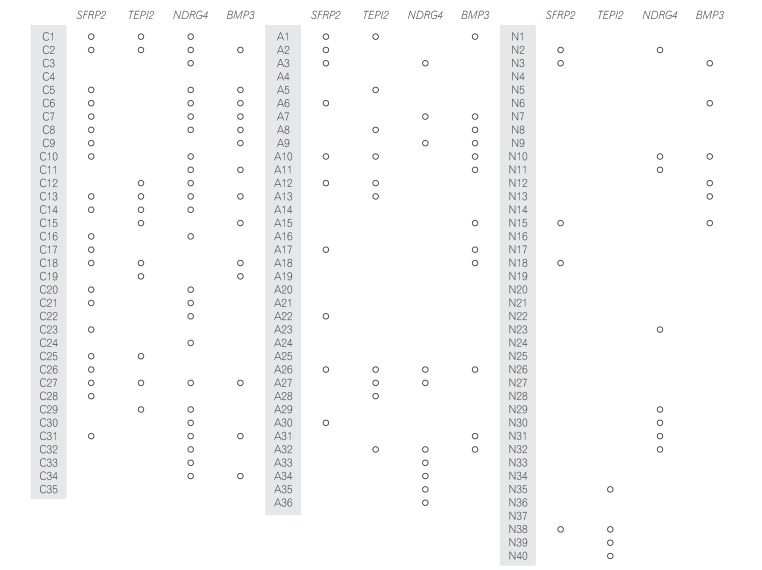
Methylated SFRP2, TFPI2, NDRG4, and BMP3 promoters were detected in 60.0%, 31.4%, 68.8%, and 40.0% of CRC samples and in 27.8%, 27.8%, 27.8%, and 33.3% of AA samples, respectively. The sensitivities obtained using 4 markers to detect CRC and AA were 94.3% and 72.2%, respectively. The specificity was 55.0%.
CONCLUSIONS
Our results demonstrate that the SFRP2, TFPI2, NDRG4, and BMP3 promoter methylation analysis of stool sample DNA showed high sensitivity but low specificity for detecting CRC and AA. Because of the low specificity, 4 methylated markers might not be sufficient for CRC screening in the Korean population. Further large-scale studies are required to validate the methylation of these markers in the Asian population and to find new markers for the Asian population.
Keyword
MeSH Terms
Figure
Cited by 1 articles
-
Novel biomarkers for the diagnosis and prognosis of colorectal cancer
Hyung-Hoon Oh, Young-Eun Joo
Intest Res. 2020;18(2):168-183. doi: 10.5217/ir.2019.00080.
Reference
-
1. Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008; 58:71–96. PMID: 18287387.
Article2. Jung KW, Park S, Kong HJ, et al. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2009. Cancer Res Treat. 2012; 44:11–24. PMID: 22500156.
Article3. Ahlquist DA, Zou H, Domanico M, et al. Next-generation stool DNA test accurately detects colorectal cancer and large adenomas. Gastroenterology. 2012; 142:248–256. PMID: 22062357.
Article4. Markowitz SD, Bertagnolli MM. Molecular origins of cancer: molecular basis of colorectal cancer. N Engl J Med. 2009; 361:2449–2460. PMID: 20018966.5. Imperiale TF, Ransohoff DF, Itzkowitz SH, Turnbull BA, Ross ME; Colorectal Cancer Study Group. Fecal DNA versus fecal occult blood for colorectal-cancer screening in an average-risk population. N Engl J Med. 2004; 351:2704–2714. PMID: 15616205.
Article6. Lansdorp-Vogelaar I, Kuntz KM, Knudsen AB, Wilschut JA, Zauber AG, van Ballegooijen M. Stool DNA testing to screen for colorectal cancer in the Medicare population: a cost-effectiveness analysis. Ann Intern Med. 2010; 153:368–377. PMID: 20855801.
Article7. Kim ER, Kim YH. Clinical application of genetics in management of colorectal cancer. Intest Res. 2014; 12:184–193. PMID: 25349592.
Article8. Nagasaka T, Tanaka N, Cullings HM, et al. Analysis of fecal DNA methylation to detect gastrointestinal neoplasia. J Natl Cancer Inst. 2009; 101:1244–1258. PMID: 19700653.
Article9. De Maio G, Rengucci C, Zoli W, Calistri D. Circulating and stool nucleic acid analysis for colorectal cancer diagnosis. World J Gastroenterol. 2014; 20:957–967. PMID: 24574768.
Article10. A stool DNA test (Cologuard) for colorectal cancer screening. JAMA. 2014; 312:2566. PMID: 25514307.11. Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014; 370:1287–1297. PMID: 24645800.
Article12. Chang E, Park DI, Kim YJ, et al. Detection of colorectal neoplasm using promoter methylation of ITGA4, SFRP2, and p16 in stool samples: a preliminary report in Korean patients. Hepatogastroenterology. 2010; 57:720–727. PMID: 21033217.13. Mandel JS, Bond JH, Church TR, et al. Reducing mortality from colorectal cancer by screening for fecal occult blood: Minnesota Colon Cancer Control study. N Engl J Med. 1993; 328:1365–1371. PMID: 8474513.
Article14. Ahlquist DA. Molecular detection of colorectal neoplasia. Gastroenterology. 2010; 138:2127–2139. PMID: 20420950.
Article15. Osborn NK, Ahlquist DA. Stool screening for colorectal cancer: molecular approaches. Gastroenterology. 2005; 128:192–206. PMID: 15633136.
Article16. Lidgard GP, Domanico MJ, Bruinsma JJ, et al. Clinical performance of an automated stool DNA assay for detection of colorectal neoplasia. Clin Gastroenterol Hepatol. 2013; 11:1313–1318. PMID: 23639600.
Article17. Itzkowitz S, Brand R, Jandorf L, et al. A simplified, noninvasive stool DNA test for colorectal cancer detection. Am J Gastroenterol. 2008; 103:2862–2870. PMID: 18759824.
Article18. Sidransky D, Tokino T, Hamilton SR, et al. Identification of ras oncogene mutations in the stool of patients with curable colorectal tumors. Science. 1992; 256:102–105. PMID: 1566048.
Article19. Fodde R, Smits R, Clevers H. APC, signal transduction and genetic instability in colorectal cancer. Nat Rev Cancer. 2001; 1:55–67. PMID: 11900252.
Article20. Xu Q, D'Amore PA, Sokol SY. Functional and biochemical interactions of Wnts with FrzA, a secreted Wnt antagonist. Development. 1998; 125:4767–4776. PMID: 9806925.
Article21. Glöckner SC, Dhir M, Yi JM, et al. Methylation of TFPI2 in stool DNA: a potential novel biomarker for the detection of colorectal cancer. Cancer Res. 2009; 69:4691–4699. PMID: 19435926.22. Melotte V, Lentjes MH, van den Bosch SM, et al. N-myc downstream-regulated gene 4 (NDRG4): a candidate tumor suppressor gene and potential biomarker for colorectal cancer. J Natl Cancer Inst. 2009; 101:916–927. PMID: 19535783.
Article23. Loh K, Chia JA, Greco S, et al. Bone morphogenic protein 3 inactivation is an early and frequent event in colorectal cancer development. Genes Chromosomes Cancer. 2008; 47:449–460. PMID: 18311777.
Article24. Park SK, Song CS, Yang HJ, et al. Field cancerization in sporadic colon cancer. Gut Liver. 2016; 10:773–780. PMID: 27114416.
Article25. US Preventive Services Task Force, Bibbins-Domingo K, Grossman DC, et al. Screening for colorectal cancer: US Preventive Services Task Force recommendation statement. JAMA. 2016; 315:2564–2575. PMID: 27304597.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Field Cancerization in Sporadic Colon Cancer
- Hypermethylated promoters of tumor suppressor genes were identified in Crohn’s disease patients
- In silico Identification of SFRP1 as a Hypermethylated Gene in Colorectal Cancers
- LUCAT1 Epigenetically Downregulates the Tumor Suppressor Genes CXXC4 and SFRP2 in Gastric Cancer
- Classification of Colon Cancer Patients Based on the Methylation Patterns of Promoters