J Bacteriol Virol.
2017 Sep;47(3):156-164. 10.4167/jbv.2017.47.3.156.
Study on Causes of Respiratory Disease to the Middle East Respiratory Syndrome-negative Subjects
- Affiliations
-
- 1Busan Metropolitan City Institute of Health & Environment, Busan, Korea. kies98@korea.kr
- KMID: 2392964
- DOI: http://doi.org/10.4167/jbv.2017.47.3.156
Abstract
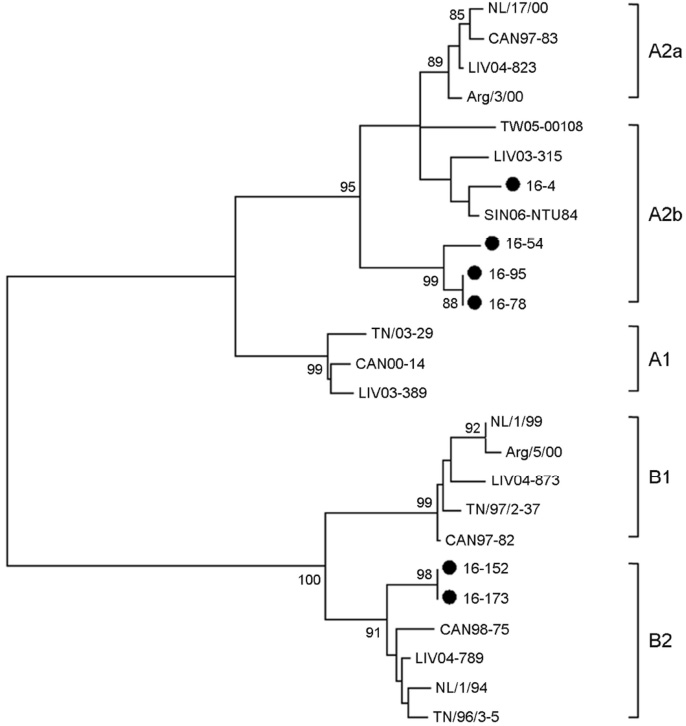
- Middle East respiratory syndrome coronavirus (MERS-CoV) causes severe cases of human respiratory disease. The current outbreak of infection with this virus in South Korea, which began on May 20, 2015, has infected 186 patients and caused 36 deaths within 2 months. In this study, to investigate the viral pathogen causing acute respiratory infections, multiplex/RT-PCR was performed on were obtained from nucleic acid of the Middle East Respiratory Syndrome-negative subjects. Viruses and atypical bacteria were detected in 39 of 337 (11.6%). Frequent viruses were human rhinovirus (n=11, 3.3%), human metapneumovirus (n=9, 2.7%), parainfluenza (n=9, 2.7%) and adenovirus (n=4, 1.2%). Mycoplasma pneumonia (M. pneumonia) was detected in 1.8 % (n=6). Out of 9 human metapneumovirus (hMPV) positive samples, 6 samples were successfully sequenced using F gene. And M. pneumoniae was sequencing of a repetitive region of the P1 gene. Phylogenetic analysis revealed that hMPV clustered into A2b lineage (n=4), B2 lineage (n=2) and M. pneumoniae clustered into two genotypes: Type 1 (n=4), Type 2a (n=2).
Keyword
MeSH Terms
Figure
Reference
-
1. World Health Organization (WHO). Middle East respiratory syndrome coronavirus (MERS-CoV) global summary and risk assessment. 2016. Available at http://who.int/emergencies/mers-cov/mers-summary-2016.pdf.2. Zumla A, Hui DS, Perlman S. Middle East respiratory syndrome. Lancet. 2015; 386:995–1007.
Article3. Reiche J, Jacobsen S, Neubauer K, Hafemann S, Nitsche A, Milde J, et al. Human metapneumo-virus: insights from a ten-year molecular and epidemiological analysis in Germany. PLoS One. 2014; 9:e88342.
Article4. Dumke R, Lück PC, Noppen C, Schaefer C, von Baum H, Marre R, et al. Culture-independent molecular subtyping of Mycoplasma pneumoniae in clinical samples. J Clin Microbiol. 2006; 44:2567–2570.
Article5. Mackay IM. Human rhinoviruses: the cold wars resume. J Clin Virol. 2008; 42:297–320.
Article6. Hayden FG. Rhinovirus and the lower respiratory tract. Rev Med Virol. 2004; 14:17–31.
Article7. Korea Centers for Disease Control and Prevention (KCDC). Analysis of pathogen in acute respiratory infections. Public Health Weekly Report. 2014; 4:65–72.8. KCDC . The prevalence of the respiratory viruses in the patients with acute respiratory infections 2014. Public Health Weekly Report. 2016; 9:26–36.9. Park JS. Acute viral lower respiratory tract infections in children. Korean J Pediatr. 2009; 52:269–276.
Article10. Kang SY, Hong CR, Kang HM, Cho EY, Lee HJ, Choi EH, et al. Clinical and epidemiological characteristics of human metapneumovirus infections in comparison with respiratory syncytial virus A and B. Korean J Pediatr Infect Dis. 2013; 20:168–177.
Article11. Williams JV, Harris PA, Tollefson SJ, Halburnt-Rush LL, Pingsterhaus JM, Edwards KM, et al. Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. N Engl J Med. 2004; 350:443–450.
Article12. van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, et al. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001; 7:719–724.
Article13. Chung JY, Han TH, Kim BE, Kim CK, Kim SW, Hwang ES. Human metapneumovirus infection in hospitalized children with acute respiratory disease in Korea. J Korean Med Sci. 2006; 21:838–842.
Article14. Yang SJ. Etiology of respiratory virus of acute respiratory infection in adult population at a single tertiary care center. Masters Thesis. Gachon University, School of Medicine;2017.15. Collins PL, Chanock RM, McIntosh K. Parainfluenza viruses. In : Fields BN, editor. Virology. Philadelphia: Lippincott-Raven;1996. p. 1205–1241.16. Kim WK, Kim KE. Mycoplasma and chlamydia infection in Korea. Korean J Pediatr. 2009; 52:277–282.17. KCDC . Analysis of genotype distribution and detection rate of Mycoplasma pneumoniae according to respiratory disease type. Public Health Weekly Report. 2014; 9:177–181.18. Falsey AR, Walsh EE. Viral pneumonia in older adults. Clin Infect Dis. 2006; 42:518–524.
Article19. Kahn JS. Epidemiology of human metapneumovirus. Clin Microbiol Rev. 2006; 19:546–557.
Article20. Huck B, Scharf G, Neumann-Haefelin D, Puppe W, Weigl J, Falcone V. Novel human metapneumovirus sublineage. Emerg Infect Dis. 2006; 12:147–150.
Article21. Neemuchwala A, Duvvuri VR, Marchand-Austin A, Li A, Gubbay JB. Human metapneumovirus prevalence and molecular epidemiology in respiratory outbreaks in Ontario, Canada. J Med Virol. 2015; 87:269–274.
Article22. Kim HR, Cho AR, Lee MK, Yun SW, Kim TH. Genotype variability and clinical features of human metapneumovirus isolated from Korean children, 2007 to 2010. J Mol Diagn. 2012; 14:61–64.
Article23. Chung JY, Han TH, Kim SW, Hwang ES. Genotype variability of human metapneumovirus, South Korea. J Med Virol. 2008; 80:902–905.
Article24. Waites KB, Talkington DF. Mycoplasm pneumoniae and its role as a human pathogen. Clin Microbiol Rev. 2004; 17:697–728.
Article25. Matsuoka M, Narita M, Okazaki N, Ohya H, Yamazaki T, Ouchi K, et al. Characterization and molecular analysis of macrolide resistance Mycoplasma pneumoniae clinical isolates obtained in Japan. Antimicrob Agents Chemother. 2004; 48:4624–4630.
Article26. Xin D, Mi Z, Han X, Qin L, Li J, Wei T, et al. Molecular mechanisms of macrolide resistance in clinical isolates of Mycoplasma pneumoniae from China. Antimicrob Agents Chemother. 2009; 53:2158–2159.
Article27. Dégrange S, Cazanave C, Charron A, Renaudin H, Bébéar C, Bébéar CM. Development of multiple-locus variable-number tandem-repeat analysis for molecular typing of Mycoplasma pneumoniae. J Clin Microbiol. 2009; 47:914–923.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- An Unexpected Outbreak of Middle East Respiratory Syndrome Coronavirus Infection in the Republic of Korea, 2015
- The Korean Middle East Respiratory Syndrome Coronavirus Outbreak and Our Responsibility to the Global Scientific Community
- The Same Middle East Respiratory Syndrome-Coronavirus (MERS-CoV) yet Different Outbreak Patterns and Public Health Impacts on the Far East Expert Opinion from the Rapid Response Team of the Republic of Korea
- Emergency cesarean section in an epidemic of the middle east respiratory syndrome: a case report
- Costly Lessons From the 2015 Middle East Respiratory Syndrome Coronavirus Outbreak in Korea