Ann Surg Treat Res.
2017 Oct;93(4):173-180. 10.4174/astr.2017.93.4.173.
Establishing a colorectal cancer liver metastasis patient-derived tumor xenograft model for the evaluation of personalized chemotherapy
- Affiliations
-
- 1College of Pharmacy, Duksung Women's University, Seoul, Korea.
- 2Innovative Drug Center, Duksung Women's University, Seoul, Korea.
- 3Department of Pathology, Gangnam Severance Hospital, Yonsei University, Seoul, Korea.
- 4Pancreatobiliary Cancer Clinic, Department of Surgery, Gangnam Severance Hospital, Yonsei University, Seoul, Korea. jspark330@yuhs.ac
- KMID: 2392366
- DOI: http://doi.org/10.4174/astr.2017.93.4.173
Abstract
- PURPOSE
In order to suggest optimal anticancer drugs for patient-tailored chemotherapy, we developed a colorectal cancer (CRC)-liver metastasis patient-derived tumor xenograft (PDTX) model.
METHODS
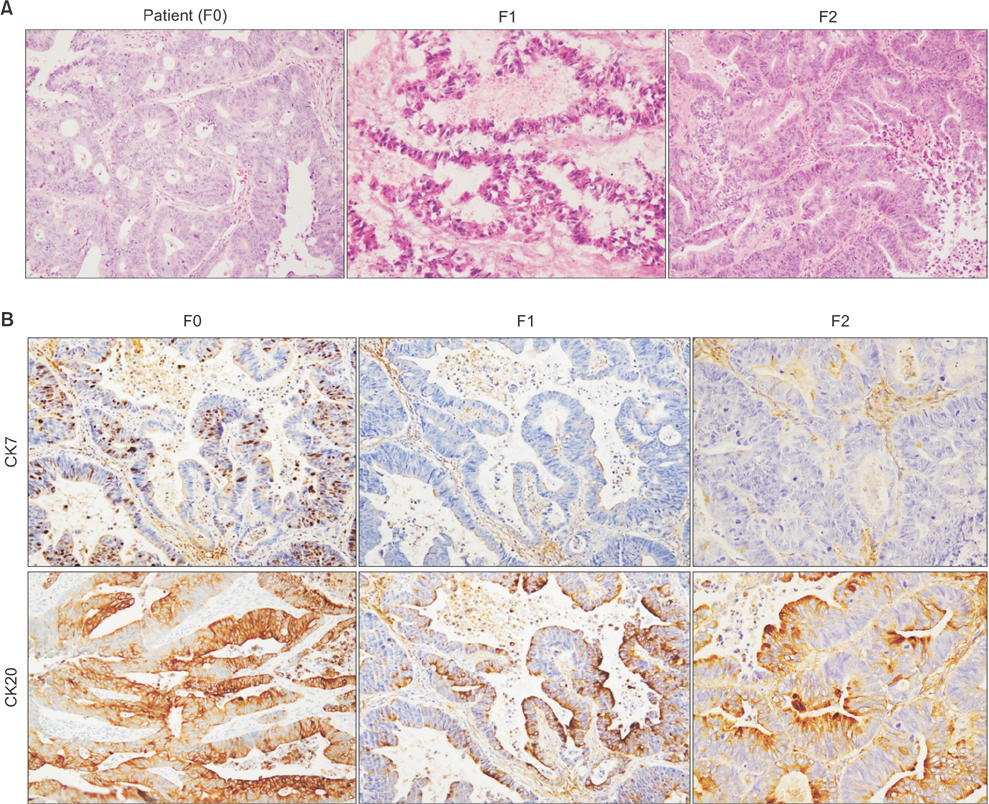
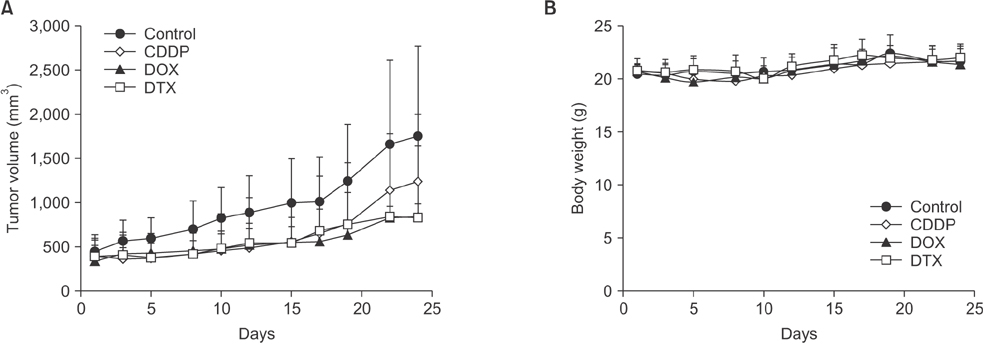
Tissue obtained from a patient with CRC-liver metastasis (F0) was transplanted in a nonobese female mouse with diabetic/severe combined immune deficiency (F1) and the tumor tissue was retransplanted into nude mice (F2). When tumor volumes reached ~500 mm³, the F2 mice were randomly divided into 4 groups (n = 4/group) of doxorubicin, cisplatin, docetaxel, and nontreated control groups. The tumor tissues were investigated using H&E staining, terminal deoxynucleotidyl transferase dUTP nick end labeling assays, and immunohistochemistry. To determine where the mutant allele frequencies varied across the different passages, we isolated genomic DNA from the primary tumor, liver metastasis, and PDTX models (F1/F2).
RESULTS
The physiological properties of the tumor were in accord with those of the patient's tumors. Anticancer drugs delayed tumor growth, inhibited proliferation, and caused apoptosis. Histological assessments revealed no observable heterogeneity among the intragenerational PDTX models. Target exon sequencing analysis without high-quality filter conditions revealed some genetic variations in the 83 cancer-related genes across the generations. However, when de novo mutations were defined as a total count of zero in F0 and ≥5 in F2, exactly prognostic impact of clone cancer profiling (EGFR, KRAS, BRAF, PIK3CA, NRAS, APC and TP53) were detected in the paired.
CONCLUSION
A CRC liver metastasis PDTX model was established for the evaluation of chemotherapeutic efficacy. This model retained the physiological characters of the patient tumors and potentially provides a powerful means of assessing chemotherapeutic efficacy.
MeSH Terms
-
Animals
Apoptosis
Cisplatin
Clone Cells
Colorectal Neoplasms*
DNA
DNA Nucleotidylexotransferase
Doxorubicin
Drug Therapy*
Exons
Family Characteristics
Female
Gene Frequency
Genetic Variation
Heterografts*
Humans
Immunohistochemistry
Liver*
Mice
Mice, Nude
Neoplasm Metastasis*
Population Characteristics
Sequence Analysis
Xenograft Model Antitumor Assays
Cisplatin
DNA
DNA Nucleotidylexotransferase
Doxorubicin
Figure
Reference
-
1. Jung KW, Won YJ, Kong HJ, Oh CM, Cho H, Lee DH, et al. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2012. Cancer Res Treat. 2015; 47:127–141.2. Puig I, Chicote I, Tenbaum SP, Arques O, Herance JR, Gispert JD, et al. A personalized preclinical model to evaluate the metastatic potential of patient-derived colon cancer initiating cells. Clin Cancer Res. 2013; 19:6787–6801.3. Bendell JC, Zakari A, Peyton JD, Boccia R, Moskowitz M, Gian V, et al. A Phase II Study of FOLFOXIRI Plus Panitumumab Followed by Evaluation for Resection in Patients With Metastatic KRAS Wild-Type Colorectal Cancer With Liver Metastases Only. Oncologist. 2016; 21:279–280.4. Akgul O, Cetinkaya E, Ersoz S, Tez M. Role of surgery in colorectal cancer liver metastases. World J Gastroenterol. 2014; 20:6113–6122.5. Whittle JR, Lewis MT, Lindeman GJ, Visvader JE. Patient-derived xenograft models of breast cancer and their predictive power. Breast Cancer Res. 2015; 17:17.6. Izumchenko E, Meir J, Bedi A, Wysocki PT, Hoque MO, Sidransky D. Patient-derived xenografts as tools in pharmaceutical development. Clin Pharmacol Ther. 2016; 99:612–621.7. Gao H, Korn JM, Ferretti S, Monahan JE, Wang Y, Singh M, et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med. 2015; 21:1318–1325.8. Cho YB, Hong HK, Choi YL, Oh E, Joo KM, Jin J, et al. Colorectal cancer patient-derived xenografted tumors maintain characteristic features of the original tumors. J Surg Res. 2014; 187:502–509.9. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760.10. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011; 43:491–498.11. Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013; 43:11.10.1–11.10.33.12. Wilm A, Aw PP, Bertrand D, Yeo GH, Ong SH, Wong CH, et al. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012; 40:11189–11201.13. Wei Z, Wang W, Hu P, Lyon GJ, Hakonarson H. SNVer: a statistical tool for variant calling in analysis of pooled or individual next-generation sequencing data. Nucleic Acids Res. 2011; 39:e132.14. Nemati F, Sastre-Garau X, Laurent C, Couturier J, Mariani P, Desjardins L, et al. Establishment and characterization of a panel of human uveal melanoma xenografts derived from primary and/or metastatic tumors. Clin Cancer Res. 2010; 16:2352–2362.15. Quintana E, Piskounova E, Shackleton M, Weinberg D, Eskiocak U, Fullen DR, et al. Human melanoma metastasis in NSG mice correlates with clinical outcome in patients. Sci Transl Med. 2012; 4:159ra149.16. Boone JD, Dobbin ZC, Straughn JM Jr, Buchsbaum DJ. Ovarian and cervical cancer patient derived xenografts: The past, present, and future. Gynecol Oncol. 2015; 138:486–491.17. Dong X, Guan J, English JC, Flint J, Yee J, Evans K, et al. Patient-derived first generation xenografts of non-small cell lung cancers: promising tools for predicting drug responses for personalized chemotherapy. Clin Cancer Res. 2010; 16:1442–1451.18. Mohseni MJ, Amanpour S, Muhammadnejad S, Sabetkish S, Muhammadnejad A, Heidari R, et al. Establishment of a patient-derived Wilms’ tumor xenograft model: a promising tool for individualized cancer therapy. J Pediatr Urol. 2014; 10:123–129.19. Zhu Y, Tian T, Li Z, Tang Z, Wang L, Wu J, et al. Establishment and characterization of patient-derived tumor xenograft using gastroscopic biopsies in gastric cancer. Sci Rep. 2015; 5:8542.20. Hao C, Wang L, Peng S, Cao M, Li H, Hu J, et al. Gene mutations in primary tumors and corresponding patient-derived xenografts derived from non-small cell lung cancer. Cancer Lett. 2015; 357:179–185.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Generation and Application of Patient-Derived Xenograft Model for Cancer Research
- Comparison of the Genetic Alterations between Primary Colorectal Cancers and Their Corresponding Patient-Derived Xenograft Tissues
- Sex-dependent liver cancer xenograft models for predicting clinical data in the evaluation of anticancer drugs
- Successful Xenograft of Endoscopic Ultrasound-Guided Fine-Needle Aspiration Specimen from Human Extrahepatic Cholangiocarcinoma into an Immunodeficient Mouse
- The Progress and Clinical Application of Breast Cancer Organoids