J Breast Cancer.
2017 Jun;20(2):150-159. 10.4048/jbc.2017.20.2.150.
Mutations of the Epidermal Growth Factor Receptor Gene in Triple-Negative Breast Cancer
- Affiliations
-
- 1Department of Pathology, Daegu Fatima Hospital, Daegu, Korea.
- 2Department of Pathology, Yeungnam University College of Medicine, Daegu, Korea. ykbae@ynu.ac.kr
- 3Department of Surgery, Yeungnam University College of Medicine, Daegu, Korea.
- KMID: 2389752
- DOI: http://doi.org/10.4048/jbc.2017.20.2.150
Abstract
- PURPOSE
Epidermal growth factor receptor (EGFR) is considered a potential therapeutic target for anti-EGFR therapy in triple-negative breast cancer (TNBC). However, the frequency of EGFR gene mutation in TNBC is low and varies with ethnicity. This study aimed to investigate the incidence of EGFR gene mutation in TNBC.
METHODS
EGFR protein expression was evaluated by immunohistochemistry in tissue microarrays of 493 TNBC cases using four different primary antibodies, which included mutation-specific antibodies. For cases with an immunoreactivity level ≥1+, we performed pyrosequencing analysis for EGFR gene mutation. A case was considered mutation-positive when its mutation frequency minus its limit of detection (LOD) was >10%. Cases with mutation frequency higher than LOD were assessed for EGFR gene mutation status using the Cobas assay and by peptide nucleic acid-mediated polymerase chain reaction (PNA-clamping).
RESULTS
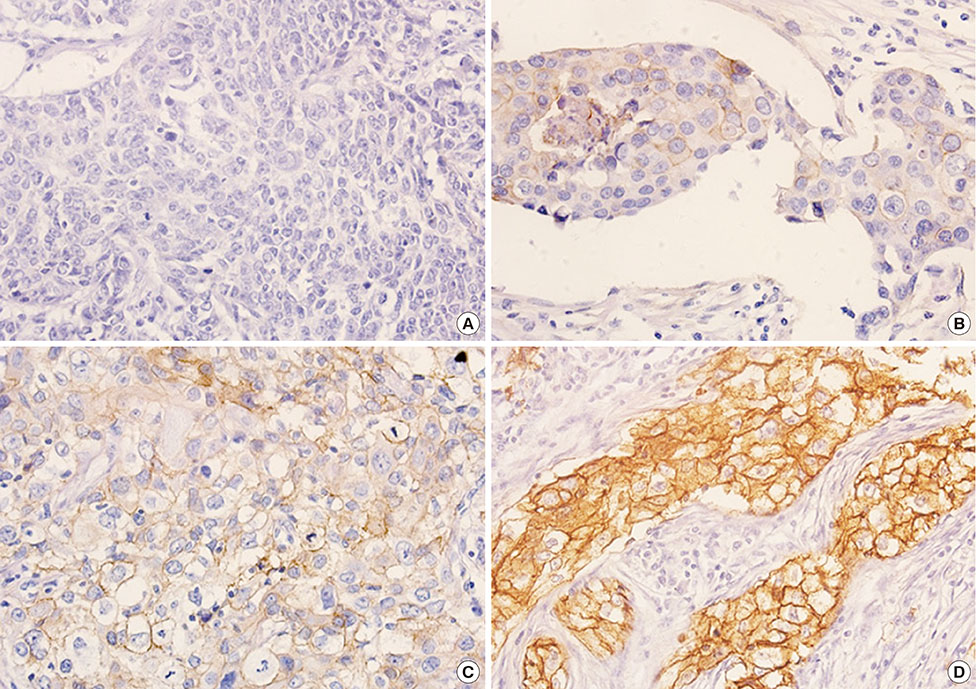
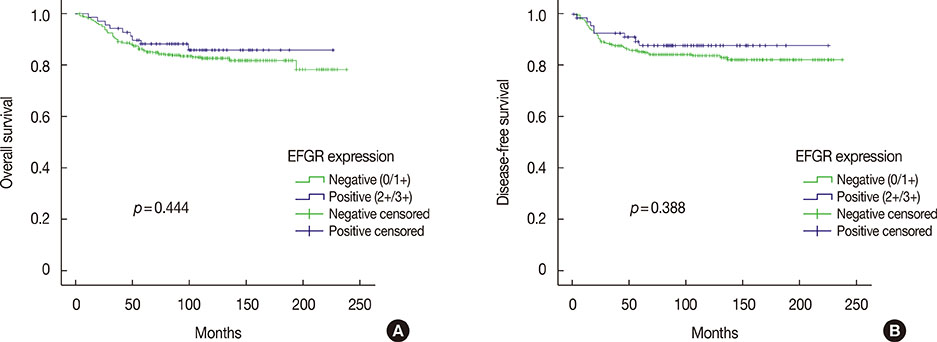
Among 493 TNBCs, 148 (30.0%) exhibited staining ≥1+ for EGFR, including 78 with 1+, 49 with 2+, and 21 with 3+. Positive EGFR expression (≥2+) was significantly associated with lymphovascular invasion (p=0.010), but not with overall survival (p=0.444) or disease-free survival (p=0.388). None of the 493 TNBCs harbored an EGFR gene mutation. Among 148 cases with an EGFR staining result ≥1+, five (3.4%) showed mutation frequencies (4.4%-10.9%) higher than LOD (2.6%-4.3%) in exons 19 (L747_P753>Q) or 21 (L858R and L861Q) as determined by pyrosequencing. However, Cobas and PNA-clamping failed to detect the presence of EGFR gene mutation in these five cases.
CONCLUSION
No activating mutation of EGFR gene of clinical significance was observed in 148 TNBC cases using three commercially available methods. Thus, EGFR gene mutation appears to be an extremely rare event in patients with TNBC.
Keyword
MeSH Terms
-
Antibodies
Breast Neoplasms
Disease-Free Survival
Epidermal Growth Factor*
Exons
Genes, erbB-1
Humans
Immunohistochemistry
Incidence
Limit of Detection
Mutation Rate
Polymerase Chain Reaction
Receptor, Epidermal Growth Factor*
Triple Negative Breast Neoplasms*
Antibodies
Epidermal Growth Factor
Receptor, Epidermal Growth Factor
Figure
Cited by 1 articles
-
Implication of Pre- and Post-radiotherapy ctDNA Dynamics in Patients with Residual Triple-Negative Breast Cancer at Surgery after Neoadjuvant Chemotherapy: Findings from a Prospective Observational Study
Tae Hoon Lee, Haeyoung Kim, Yeon Jeong Kim, Woong-Yang Park, Won Park, Won Kyung Cho, Nalee Kim
Cancer Res Treat. 2024;56(2):531-537. doi: 10.4143/crt.2023.996.
Reference
-
1. Nakai K, Hung MC, Yamaguchi H. A perspective on anti-EGFR therapies targeting triple-negative breast cancer. Am J Cancer Res. 2016; 6:1609–1623.2. Kim MJ, Ro JY, Ahn SH, Kim HH, Kim SB, Gong G. Clinicopathologic significance of the basal-like subtype of breast cancer: a comparison with hormone receptor and HER2/neu-overexpressing phenotypes. Hum Pathol. 2006; 37:1217–1226.
Article3. Masuda H, Zhang D, Bartholomeusz C, Doihara H, Hortobagyi GN, Ueno NT. Role of epidermal growth factor receptor in breast cancer. Breast Cancer Res Treat. 2012; 136:331–345.
Article4. Scagliotti GV, Selvaggi G, Novello S, Hirsch FR. The biology of epidermal growth factor receptor in lung cancer. Clin Cancer Res. 2004; 10(12 Pt 2):4227s–4232s.
Article5. Chung KY, Shia J, Kemeny NE, Shah M, Schwartz GK, Tse A, et al. Cetuximab shows activity in colorectal cancer patients with tumors that do not express the epidermal growth factor receptor by immunohistochemistry. J Clin Oncol. 2005; 23:1803–1810.
Article6. Fuller GN, Bigner SH. Amplified cellular oncogenes in neoplasms of the human central nervous system. Mutat Res. 1992; 276:299–306.
Article7. Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001; 344:783–792.
Article8. Cheng L, Alexander RE, Maclennan GT, Cummings OW, Montironi R, Lopez-Beltran A, et al. Molecular pathology of lung cancer: key to personalized medicine. Mod Pathol. 2012; 25:347–369.
Article9. Nakajima H, Ishikawa Y, Furuya M, Sano T, Ohno Y, Horiguchi J, et al. Protein expression, gene amplification, and mutational analysis of EGFR in triple-negative breast cancer. Breast Cancer. 2014; 21:66–74.
Article10. Grob TJ, Heilenkötter U, Geist S, Paluchowski P, Wilke C, Jaenicke F, et al. Rare oncogenic mutations of predictive markers for targeted therapy in triple-negative breast cancer. Breast Cancer Res Treat. 2012; 134:561–567.
Article11. Martin V, Botta F, Zanellato E, Molinari F, Crippa S, Mazzucchelli L, et al. Molecular characterization of EGFR and EGFR-downstream pathways in triple negative breast carcinomas with basal like features. Histol Histopathol. 2012; 27:785–792.12. Secq V, Villeret J, Fina F, Carmassi M, Carcopino X, Garcia S, et al. Triple negative breast carcinoma EGFR amplification is not associated with EGFR, Kras or ALK mutations. Br J Cancer. 2014; 110:1045–1052.
Article13. Toyama T, Yamashita H, Kondo N, Okuda K, Takahashi S, Sasaki H, et al. Frequently increased epidermal growth factor receptor (EGFR) copy numbers and decreased BRCA1 mRNA expression in Japanese triple-negative breast cancers. BMC Cancer. 2008; 8:309.
Article14. Tilch E, Seidens T, Cocciardi S, Reid LE, Byrne D, Simpson PT, et al. Mutations in EGFR, BRAF and RAS are rare in triple-negative and basal-like breast cancers from Caucasian women. Breast Cancer Res Treat. 2014; 143:385–392.
Article15. Jacot W, Lopez-Crapez E, Thezenas S, Senal R, Fina F, Bibeau F, et al. Lack of EGFR-activating mutations in European patients with triple-negative breast cancer could emphasise geographic and ethnic variations in breast cancer mutation profiles. Breast Cancer Res. 2011; 13:R133.
Article16. Reis-Filho JS, Pinheiro C, Lambros MB, Milanezi F, Carvalho S, Savage K, et al. EGFR amplification and lack of activating mutations in metaplastic breast carcinomas. J Pathol. 2006; 209:445–453.
Article17. Santarpia L, Qi Y, Stemke-Hale K, Wang B, Young EJ, Booser DJ, et al. Mutation profiling identifies numerous rare drug targets and distinct mutation patterns in different clinical subtypes of breast cancers. Breast Cancer Res Treat. 2012; 134:333–343.
Article18. Lv N, Xie X, Ge Q, Lin S, Wang X, Kong Y, et al. Epidermal growth factor receptor in breast carcinoma: association between gene copy number and mutations. Diagn Pathol. 2011; 6:118.
Article19. Teng YH, Tan WJ, Thike AA, Cheok PY, Tse GM, Wong NS, et al. Mutations in the epidermal growth factor receptor (EGFR) gene in triple negative breast cancer: possible implications for targeted therapy. Breast Cancer Res. 2011; 13:R35.
Article20. Kim Y, Kim J, Lee HD, Jeong J, Lee W, Lee KA. Spectrum of EGFR gene copy number changes and KRAS gene mutation status in Korean triple negative breast cancer patients. PLoS One. 2013; 8:e79014.
Article21. Park HS, Jang MH, Kim EJ, Kim HJ, Lee HJ, Kim YJ, et al. High EGFR gene copy number predicts poor outcome in triple-negative breast cancer. Mod Pathol. 2014; 27:1212–1222.
Article22. Choi JE, Kang SH, Lee SJ, Bae YK. Androgen receptor expression predicts decreased survival in early stage triple-negative breast cancer. Ann Surg Oncol. 2015; 22:82–89.
Article23. Lee HJ, Xu X, Kim H, Jin Y, Sun P, Kim JE, et al. Comparison of direct sequencing, PNA clamping-real time polymerase chain reaction, and pyrosequencing methods for the detection of EGFR mutations in non-small cell lung carcinoma and the correlation with clinical responses to EGFR tyrosine kinase inhibitor treatment. Korean J Pathol. 2013; 47:52–60.
Article24. Yoon SH, Choi YD, Oh IJ, Kim KS, Choi H, Chang J, et al. Peptide nucleic acid clamping versus direct sequencing for the detection of EGFR gene mutation in patients with non-small cell lung cancer. Cancer Res Treat. 2015; 47:661–669.
Article25. Dufort S, Richard MJ, Lantuejoul S, de Fraipont F. Pyrosequencing, a method approved to detect the two major EGFR mutations for anti EGFR therapy in NSCLC. J Exp Clin Cancer Res. 2011; 30:57.
Article26. Li AR, Chitale D, Riely GJ, Pao W, Miller VA, Zakowski MF, et al. EGFR mutations in lung adenocarcinomas: clinical testing experience and relationship to EGFR gene copy number and immunohistochemical expression. J Mol Diagn. 2008; 10:242–248.27. Brevet M, Arcila M, Ladanyi M. Assessment of EGFR mutation status in lung adenocarcinoma by immunohistochemistry using antibodies specific to the two major forms of mutant EGFR. J Mol Diagn. 2010; 12:169–176.
Article28. Kim CH, Kim SH, Park SY, Yoo J, Kim SK, Kim HK. Identification of EGFR mutations by immunohistochemistry with EGFR mutation-specific antibodies in biopsy and resection specimens from pulmonary adenocarcinoma. Cancer Res Treat. 2015; 47:653–660.
Article29. Wen YH, Brogi E, Hasanovic A, Ladanyi M, Soslow RA, Chitale D, et al. Immunohistochemical staining with EGFR mutation-specific antibodies: high specificity as a diagnostic marker for lung adenocarcinoma. Mod Pathol. 2013; 26:1197–1203.
Article30. Lamy PJ, Jacot W. Worldwide variations in EGFR somatic mutations: a challenge for personalized medicine. Diagn Pathol. 2012; 7:13.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Amplification of epidermal growth factor receptor gene in primary cervical cancer
- Amplification of epidermal growth factor receptor gene in primary cervical cancer
- MRI Findings of Triple Negative Breast Cancer: A Comparison with Non-Triple Negative Breast Cancer
- Comparative Study of Metaplastic Breast Carcinoma and Triple-Negative Breast Carcinoma Using Histologic and Immunohistochemical Analyses
- Current Issues and Clinical Evidence in Tumor-Infiltrating Lymphocytes in Breast Cancer