Cancer Res Treat.
2017 Jul;49(3):627-634. 10.4143/crt.2016.292.
Clinically Significant Unclassified Variants in BRCA1 and BRCA2 genes among Korean Breast Cancer Patients
- Affiliations
-
- 1Center for Breast Cancer, Hospital, National Cancer Center, Goyang, Korea. eslee@ncc.re.kr
- 2College of Veterinary Medicine, Konkuk University, Seoul, Korea.
- 3Department of Cancer Control and Policy, Graduate School of Cancer Science and Policy, National Cancer Center, Goyang, Korea.
- 4Biomolecular Function Research Branch, National Cancer Center, Goyang, Korea.
- 5Genetic Counseling Clinic, Hospital, National Cancer Center, Goyang, Korea. ksy@ncc.re.kr
- 6Department of System Cancer Science, Graduate School of Cancer Science and Policy, National Cancer Center, Goyang, Korea.
- 7Translational Epidemiology Branch, National Cancer Center, Goyang, Korea.
- 8Precision Medicine Branch, Research Institute, National Cancer Center, Goyang, Korea.
- KMID: 2388308
- DOI: http://doi.org/10.4143/crt.2016.292
Abstract
- PURPOSE
Unclassified variants (UVs) of BRCA1 and BRCA2 genes are not defined as pathogenic for breast cancer, and their clinical significance currently remains undefined. Therefore, this study was conducted to identify potentially pathogenic UVs by comparing their prevalence between breast cancer patients and controls.
MATERIALS AND METHODS
A total of 328 breast cancer patients underwent BRCA1/2 genetic screening at the National Cancer Center of Korea. Genetic variants of BRCA genes that were categorized as unclassified according to the Breast Cancer Information Core database were selected based on allelic frequency, after which candidate variants were genotyped in 421 healthy controls. We also examined family members of the study participants. Finally, the effects of amino acid substitutions on protein structure and function were predicted in silico.
RESULTS
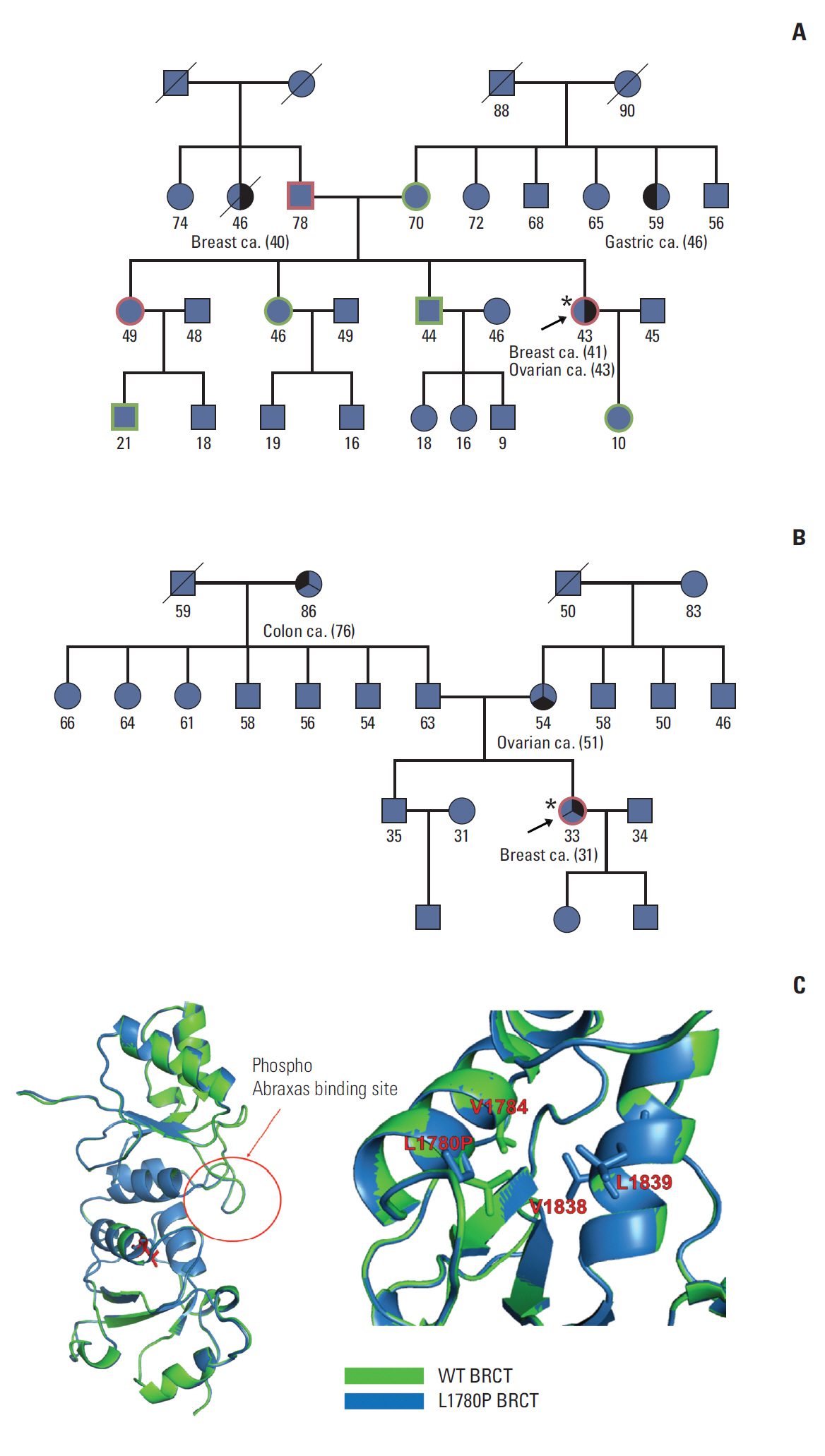
Genetic tests revealed 33 UVs in BRCA1 and 47 in BRCA2. Among 15 candidates genotyped in healthy controls, c.5339T>C in BRCA1 and c.6029T>G, c.7522G>A in BRCA2 were not detected. Moreover, the c.5339T>C variant in the BRCA1 gene was detected in four patients with a family history of breast cancer. This nonsynonymous variant (Leu1780Pro) in the BRCA1 C-terminal domain was predicted to have an effect on BRCA1 protein structure/function.
CONCLUSION
This study showed that comparison of genotype frequency between cases and controls could help identify UVs of BRCA genes that are potentially pathogenic. Moreover, ourfindings suggest that c.5339T>C in BRCA1 might be a pathogenic variant for patients and their families.
Keyword
MeSH Terms
Figure
Cited by 1 articles
-
Clinicopathological Features of Patients with the
BRCA1 c.5339T>C (p.Leu1780Pro) Variant
Hyung Seok Park, Jai Min Ryu, Ji Soo Park, Seock-Ah Im, So-Youn Jung, Eun-Kyu Kim, Woo-Chan Park, Jun Won Min, Jeeyeon Lee, Ji Young You, Jeong Eon Lee, Sung-Won Kim
Cancer Res Treat. 2020;52(3):680-688. doi: 10.4143/crt.2019.351.
Reference
-
References
1. Jung KW, Won YJ, Kong HJ, Oh CM, Cho H, Lee DH, et al. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2012. Cancer Res Treat. 2015; 47:127–41.
Article2. Futreal PA, Liu Q, Shattuck-Eidens D, Cochran C, Harshman K, Tavtigian S, et al. BRCA1 mutations in primary breast and ovarian carcinomas. Science. 1994; 266:120–2.3. Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994; 266:66–71.4. Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995; 378:789–92.
Article5. Son BH, Ahn SH, Kim SW, Kang E, Park SK, Lee MH, et al. Prevalence of BRCA1 and BRCA2 mutations in non-familial breast cancer patients with high risks in Korea: the Korean Hereditary Breast Cancer (KOHBRA) Study. Breast Cancer Res Treat. 2012; 133:1143–52.
Article6. Kang E, Seong MW, Park SK, Lee JW, Lee J, Kim LS, et al. The prevalence and spectrum of BRCA1 and BRCA2 mutations in Korean population: recent update of the Korean Hereditary Breast Cancer (KOHBRA) study. Breast Cancer Res Treat. 2015; 151:157–68.
Article7. Szabo C, Masiello A, Ryan JF, Brody LC. The breast cancer information core: database design, structure, and scope. Hum Mutat. 2000; 16:123–31.
Article8. Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014; 42:D980–5.
Article9. Harrison SM, Riggs ER, Maglott DR, Lee JM, Azzariti DR, Niehaus A, et al. Using ClinVar as a resource to support variant interpretation. Curr Protoc Hum Genet. 2016; 89:8.
Article10. Easton DF, Deffenbaugh AM, Pruss D, Frye C, Wenstrup RJ, Allen-Brady K, et al. A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am J Hum Genet. 2007; 81:873–83.
Article11. Calo V, Bruno L, La Paglia L, Perez M, Margarese N, Di Gaudio F, et al. The clinical significance of unknown sequence variants in BRCA genes. Cancers (Basel). 2010; 2:1644–60.12. Spearman AD, Sweet K, Zhou XP, McLennan J, Couch FJ, Toland AE. Clinically applicable models to characterize BRCA1 and BRCA2 variants of uncertain significance. J Clin Oncol. 2008; 26:5393–400.
Article13. Gomez Garcia EB, Oosterwijk JC, Timmermans M, van Asperen CJ, Hogervorst FB, Hoogerbrugge N, et al. A method to assess the clinical significance of unclassified variants in the BRCA1 and BRCA2 genes based on cancer family history. Breast Cancer Res. 2009; 11:R8.
Article14. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010; 7:248–9.
Article15. Flanagan SE, Patch AM, Ellard S. Using SIFT and PolyPhen to predict loss-of-function and gain-of-function mutations. Genet Test Mol Biomarkers. 2010; 14:533–7.
Article16. Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013; Chapter 7:Unit7.
Article17. Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012; 40:W452–7.
Article18. Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014; 42:W252–8.
Article19. Glover JN. Insights into the molecular basis of human hereditary breast cancer from studies of the BRCA1 BRCT domain. Fam Cancer. 2006; 5:89–93.
Article20. Campbell SJ, Edwards RA, Glover JN. Comparison of the structures and peptide binding specificities of the BRCT domains of MDC1 and BRCA1. Structure. 2010; 18:167–76.
Article21. Coquelle N, Green R, Glover JN. Impact of BRCA1 BRCT domain missense substitutions on phosphopeptide recognition. Biochemistry. 2011; 50:4579–89.
Article22. Zhang Y, Long J, Lu W, Shu XO, Cai Q, Zheng Y, et al. Rare coding variants and breast cancer risk: evaluation of susceptibility Loci identified in genome-wide association studies. Cancer Epidemiol Biomarkers Prev. 2014; 23:622–8.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Novel Germline Mutations of BRCA1 and BRCA2 in Korean Familial Breast Cancer Patients
- Distribution of BRCA1 and BRCA2 Mutations in Asian Patients with Breast Cancer
- Frequency of BRCA1 and BRCA2 Germline Mutations Detected by Protein Truncation Test and Cumulative Risks of Breast and Ovarian Cancer among Mutation Carriers in Japanese Breast Cancer Families
- Clinicopathological Characterization of Double Heterozygosity for BRCA1 and BRCA2 Variants in Korean Breast Cancer Patients
- BRCA1/BRCA2 Pathogenic Variant Breast Cancer: Treatment and Prevention Strategies