Genetic Profiles of Korean Patients With Glucose-6-Phosphate Dehydrogenase Deficiency
- Affiliations
-
- 1Department of Laboratory Medicine, College of Medicine, The Catholic University of Korea, Seoul, Korea. miziro@catholic.ac.kr
- 2Catholic Genetic Laboratory Center, College of Medicine, The Catholic University of Korea, Seoul, Korea.
- 3Department of Pediatrics, College of Medicine, The Catholic University of Korea, Seoul, Korea. chobinkr@catholic.ac.kr
- KMID: 2373627
- DOI: http://doi.org/10.3343/alm.2017.37.2.108
Abstract
- BACKGROUND
We describe the genetic profiles of Korean patients with glucose-6-phosphate dehydrogenase (G6PD) deficiencies and the effects of G6PD mutations on protein stability and enzyme activity on the basis of in silico analysis.
METHODS
In parallel with a genetic analysis, the pathogenicity of G6PD mutations detected in Korean patients was predicted in silico. The simulated effects of G6PD mutations were compared to the WHO classes based on G6PD enzyme activity. Four previously reported mutations and three newly diagnosed patients with missense mutations were estimated.
RESULTS
One novel mutation (p.Cys385Gly, labeled G6PD Kangnam) and two known mutations [p.Ile220Met (G6PD São Paulo) and p.Glu416Lys (G6PD Tokyo)] were identified in this study. G6PD mutations identified in Koreans were also found in Brazil (G6PD São Paulo), Poland (G6PD Seoul), United States of America (G6PD Riley), Mexico (G6PD Guadalajara), and Japan (G6PD Tokyo). Several mutations occurred at the same nucleotide, but resulted in different amino acid residue changes in different ethnic populations (p.Ile380 variant, G6PD Calvo Mackenna; p.Cys385 variants, Tomah, Madrid, Lynwood; p.Arg387 variant, Beverly Hills; p.Pro396 variant, Bari; and p.Pro396Ala in India). On the basis of the in silico analysis, Class I or II mutations were predicted to be highly deleterious, and the effects of one Class IV mutation were equivocal.
CONCLUSIONS
The genetic profiles of Korean individuals with G6PD mutations indicated that the same mutations may have arisen by independent mutational events, and were not derived from shared ancestral mutations. The in silico analysis provided insight into the role of G6PD mutations in enzyme function and stability.
MeSH Terms
-
Asian Continental Ancestry Group/*genetics
Child
Child, Preschool
DNA/chemical synthesis/genetics/metabolism
Exons
Glucosephosphate Dehydrogenase/chemistry/*genetics/metabolism
Glucosephosphate Dehydrogenase Deficiency/*genetics/pathology
Humans
Male
Mutation, Missense
Polymorphism, Genetic
Protein Structure, Tertiary
Republic of Korea
Sequence Analysis, DNA
DNA
Glucosephosphate Dehydrogenase
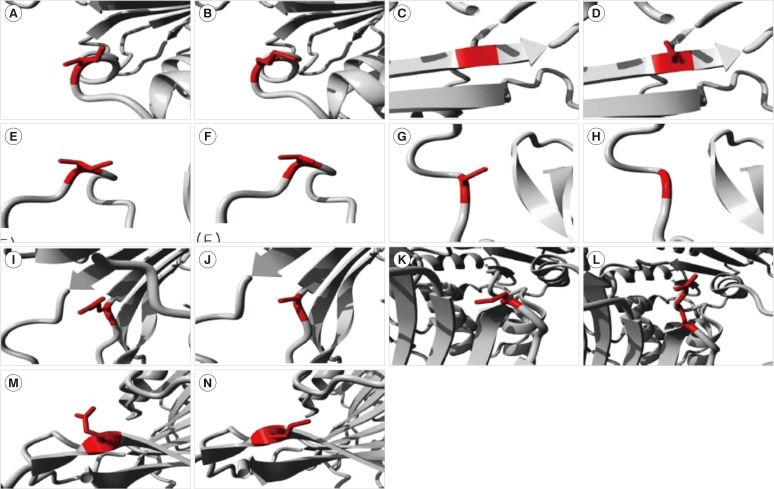
Figure
Cited by 2 articles
-
Epidemiological Study of Hereditary Hemolytic Anemia in the Korean Pediatric Population during 1997–2016: a Nationwide Retrospective Cohort Study
Ye Jee Shim, Hye Lim Jung, Hee Young Shin, Hyoung Jin Kang, Jung Yoon Choi, Jeong Ok Hah, Jae Min Lee, Young Tak Lim, Eu Jeen Yang, Hee Jo Baek, Hyoung Soo Choi, Keon Hee Yoo, Jun Eun Park, Seongkoo Kim, Ji Yoon Kim, Eun Sil Park, Ho Joon Im, Hee Won Chueh, Soon Ki Kim, Jae Hee Lee, Eun Sun Yoo, Hyeon Jin Park, Jun Ah Lee, Meerim Park, Hyun Sik Kang, Ji Kyoung Park, Na Hee Lee, Sang Kyu Park, Young-Ho Lee, Seong Wook Lee, Eun Jin Choi, Seom Gim Kong,
J Korean Med Sci. 2020;35(33):e279. doi: 10.3346/jkms.2020.35.e279.Diagnostic approaches for inherited hemolytic anemia in the genetic era
Yonggoo Kim, Joonhong Park, Myungshin Kim
Blood Res. 2017;52(2):84-94. doi: 10.5045/br.2017.52.2.84.
Reference
-
1. Cappellini MD, Fiorelli G. Glucose-6-phosphate dehydrogenase deficiency. Lancet. 2008; 371:64–74. PMID: 18177777.2. Glucose-6-phosphate dehydrogenase deficiency. WHO Working Group. Bull World Health Organ. 1989; 67:601–611. PMID: 2633878.3. Park SW, Lee HJ, Lee W, Whang KT. A Case of Glucose-6-phosphate dehydrogenase Riley Causing Hemolytic Anemia. Korean J Hematol. 1999; 34:334–337.4. Lee GB, Lee SJ, Kim YJ, Kim SY, Kim HH, Cho B, et al. A Case of G-6-PD Guadalajara. Korean J Pediatr. 2004; 47:210–213.5. Beutler E, Vulliamy TJ. Hematologically important mutations: glucose-6-phosphate dehydrogenase. Blood Cells Mol Dis. 2002; 28:93–103. PMID: 12064901.6. Jang MA, Kim J-Y, Lee K-O, Kim S-H, Koo HH, Kim H-J. A Novel de novo Mutation in the G6PD Gene in a Korean Boy with Glucose-6-phosphate Dehydrogenase Deficiency: Case Report. Ann Clin Lab Sci. 2015; 45:446–448. PMID: 26275698.7. Stenson P, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014; 133:1–9. PMID: 24077912.8. Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014; 42:D980–D985. PMID: 24234437.9. Xue Y, Chen Y, Ayub Q, Huang N, Ball EV, Mort M, et al. Deleterious- and Disease-Allele Prevalence in Healthy Individuals: Insights from Current Predictions, Mutation Databases, and Population-Scale Resequencing. Am J Hum Genet. 2012; 91:1022–1032. PMID: 23217326.10. Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012; 40:W452–W457. PMID: 22689647.11. Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015; 31:2745–2747. PMID: 25851949.12. Stone EA, Sidow A. Physicochemical constraint violation by missense substitutions mediates impairment of protein function and disease severity. Genome Res. 2005; 15:978–986. PMID: 15965030.13. Tavtigian SV, Byrnes GB, Goldgar DE, Thomas A. Classification of rare missense substitutions, using risk surfaces, with genetic-and molecular-epidemiology applications. Hum Mutat. 2008; 29:1342–1354. PMID: 18951461.14. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010; 7:248–249. PMID: 20354512.15. Li B, Krishnan VG, Mort ME, Xin F, Kamati KK, Cooper DN, et al. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics. 2009; 25:2744–2750. PMID: 19734154.16. De Baets G, Van Durme J, Reumers J, Maurer-Stroh S, Vanhee P, Dopazo J, et al. SNPeffect 4.0: on-line prediction of molecular and structural effects of protein-coding variants. Nucleic Acids Res. 2012; 40:D935–D939. PMID: 22075996.17. Valdar WS. Scoring residue conservation. Proteins. 2002; 48:227–241. PMID: 12112692.18. Oliveira RA, Oshiro M, Hirata MH, Hirata RD, Ribeiro GS, Medeiros TM, et al. A novel point mutation in a class IV glucose-6-phosphate dehydrogenase variant (G6PD São Paulo) and polymorphic G6PD variants in São Paulo State, Brazil. Genet Mol Biol. 2009; 32:251–254. PMID: 21637675.19. Hirono A, Fujii H, Hirono K, Kanno H, Miwa S. Molecular abnormality of a Japanese glucose-6-phosphate dehydrogenase variant (G6PD Tokyo) associated with hereditary non-spherocytic hemolytic anemia. Hum Genet. 1992; 88:347–348. PMID: 1733837.20. Beutler E, Westwood B, Prchal J, Vaca G, Bartsocas CS, Baronciani L. New glucose-6-phosphate dehydrogenase mutations from various ethnic groups. Blood. 1992; 80:255–256. PMID: 1611091.21. Beutler E, Westwood B, Melemed A, Dal Borgo P, Margolis D. Three new exon 10 glucose-6-phosphate dehydrogenase mutations. Blood Cells Mol Dis. 1995; 21:64–72. PMID: 7655862.22. Hirono A, Kuhl W, Gelbart T, Forman L, Fairbanks VF, Beutler E. Identification of the binding domain for NADP+ of human glucose-6-phosphate dehydrogenase by sequence analysis of mutants. Proc Natl Acad Sci U S A. 1989; 86:10015–10017. PMID: 2602358.23. Zarza R, Pujades A, Rovira A, Saavedra R, Fernandez J, Aymerich M, et al. Two new mutations of the glucose-6-phospate dehydrogenase (G6PD) gene associated with haemolytic anaemia: clinical, biochemical and molecular relationships. Br J Haematol. 1997; 98:578–582. PMID: 9332310.24. Bulliamy T, Luzzatto L, Hirono A, Beutler E. Hematologically important mutations: glucose-6-phosphate dehydrogenase. Blood Cells Mol Dis. 1997; 23:302–313. PMID: 9410474.25. Minucci A, Moradkhani K, Hwang MJ, Zuppi C, Giardina B, Capoluongo E. Glucose-6-phosphate dehydrogenase (G6PD) mutations database: review of the “old” and update of the new mutations. Blood Cells Mol Dis. 2012; 48:154–165. PMID: 22293322.26. Kaczorowska-Hac B, Burzynska B, Plochocka D, Zak-Jasinska K, Rawa K, Adamkiewicz-Drozynska E. The first reported case of G6PD deficiency due to Seoul mutation in Poland. Ann Hematol. 2014; 93:879–880. PMID: 24022758.27. Edison ES, Melinkeri SR, Chandy M. A novel missense mutation in glucose-6-phosphate dehydrogenase gene causing chronic nonspherocytic hemolytic anemia in an Indian family. Ann Hematol. 2006; 85:879–880. PMID: 16944148.28. Filosa S, Cai W, Galanello R, Cao A, De Mattia D, Schettini F, et al. A novel single-base mutation in the glucose 6-phosphate dehydrogenase gene is associated with chronic non-spherocytic haemolytic anaemia. Hum Genet. 1994; 94:560–562. PMID: 7959695.29. Mason PJ, Bautista JM, Gilsanz F. G6PD deficiency: the genotype-phenotype association. Blood Rev. 2007; 21:267–283. PMID: 17611006.30. Weimer TA, Salzano FM, Westwood B, Beutler E. Molecular characterization of glucose-6-phosphate dehydrogenase variants from Brazil. Hum Biol. 1993; 41–47. PMID: 8436389.31. Sukumar S, Mukherjee MB, Colah RB, Mohanty D. Molecular characterization of G6PD Insuli—a novel 989 CGC → CAC (330 Arg → His) mutation in the Indian population. Blood Cells Mol Dis. 2003; 30:246–247. PMID: 12737940.32. Vaca G, Arámbula E, Monsalvo A, Medina C, Nuñez C, Sandoval L, et al. Glucose-6-phosphate dehydrogenase (G-6-PD) mutations in Mexico: four new G-6-PD variants. Blood Cells Mol Dis. 2003; 31:112–120. PMID: 12850494.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Jaundice and Hemolytic Anemia Appearing within the First 24 Hour of Life due to Glucose-6-Phosphate Dehydrogenase Deficiency
- Glucose-6-phosphate dehydrogenase deficiency: report of 4 cases
- A Case of Glucose-6-Phosphate Dethydrogenase Deficiency with Chronic Hepatitis B
- Anesthetic management of a patient with glucose-6-phosphate dehydrogenase deficiency undergoing robot-assisted laparoscopic surgery: A case report
- Glucose-6-phosphate Dehydrogenase Deficiency