Yonsei Med J.
2015 Jul;56(4):877-886. 10.3349/ymj.2015.56.4.877.
Gene-Environment Interactions in Asthma: Genetic and Epigenetic Effects
- Affiliations
-
- 1Department of Interdisciplinary Program in Biomedical Science Major, Soonchunhyang Graduate School, Bucheon, Korea.
- 2Genome Research Center and Division of Allergy and Respiratory Medicine, Soonchunhyang University Bucheon Hospital, Bucheon, Korea. mdcspark@daum.net
- KMID: 2366325
- DOI: http://doi.org/10.3349/ymj.2015.56.4.877
Abstract
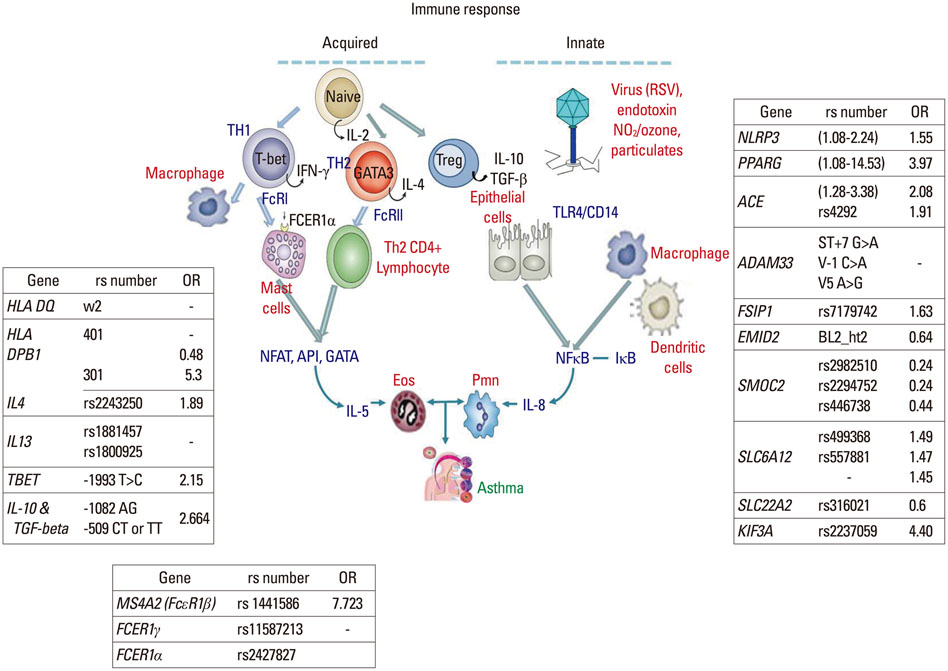
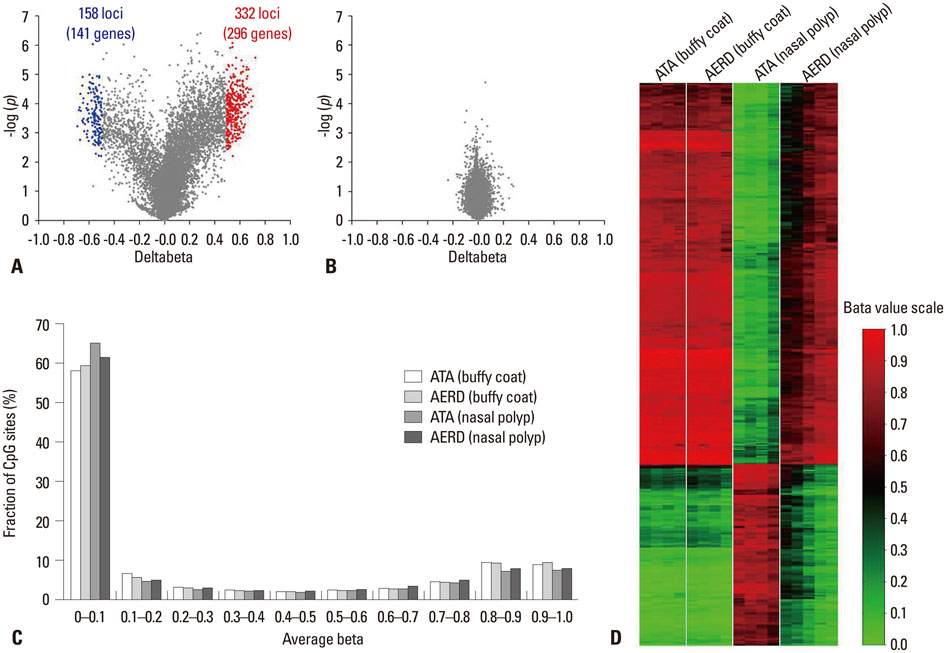
- Over the past three decades, a large number of genetic studies have been aimed at finding genetic variants associated with the risk of asthma, applying various genetic and genomic approaches including linkage analysis, candidate gene polymorphism studies, and genome-wide association studies (GWAS). However, contrary to general expectation, even single nucleotide polymorphisms (SNPs) discovered by GWAS failed to fully explain the heritability of asthma. Thus, application of rare allele polymorphisms in well defined phenotypes and clarification of environmental factors have been suggested to overcome the problem of 'missing' heritability. Such factors include allergens, cigarette smoke, air pollutants, and infectious agents during pre- and post-natal periods. The first and simplest interaction between a gene and the environment is a candidate interaction of both a well known gene and environmental factor in a direct physical or chemical interaction such as between CD14 and endotoxin or between HLA and allergens. Several GWAS have found environmental interactions with occupational asthma, aspirin exacerbated respiratory disease, tobacco smoke-related airway dysfunction, and farm-related atopic diseases. As one of the mechanisms behind gene-environment interaction is epigenetics, a few studies on DNA CpG methylation have been reported on subphenotypes of asthma, pitching the exciting idea that it may be possible to intervene at the junction between the genome and the environment. Epigenetic studies are starting to include data from clinical samples, which will make them another powerful tool for research on gene-environment interactions in asthma.
Keyword
MeSH Terms
Figure
Cited by 2 articles
-
A Six-Year Study on the Changes in Airborne Pollen Counts and Skin Positivity Rates in Korea: 2008–2013
Hye Jung Park, Jae-Hyun Lee, Kyung Hee Park, Kyu Rang Kim, Mae Ja Han, Hosoeng Choe, Jae-Won Oh, Chein-Soo Hong
Yonsei Med J. 2016;57(3):714-720. doi: 10.3349/ymj.2016.57.3.714.Near-Road Exposure and Impact of Air Pollution on Allergic Diseases in Elementary School Children: A Cross-Sectional Study
Ho Hyun Kim, Chung Soo Lee, Seung Do Yu, Jung Sub Lee, Jun Young Chang, Jun Min Jeon, Hye Rim Son, Chan Jung Park, Dong Chun Shin, Young Wook Lim
Yonsei Med J. 2016;57(3):698-713. doi: 10.3349/ymj.2016.57.3.698.
Reference
-
1. Burrows B, Martinez FD, Halonen M, Barbee RA, Cline MG. Association of asthma with serum IgE levels and skin-test reactivity to allergens. N Engl J Med. 1989; 320:271–277.
Article2. Burrows B, Sears MR, Flannery EM, Herbison GP, Holdaway MD, Silva PA. Relation of the course of bronchial responsiveness from age 9 to age 15 to allergy. Am J Respir Crit Care Med. 1995; 152(4 Pt 1):1302–1308.
Article3. Bousquet J, Chanez P, Vignola AM, Lacoste JY, Michel FB. Eosinophil inflammation in asthma. Am J Respir Crit Care Med. 1994; 150(5 Pt 2):S33–S38.
Article4. Edfors-Lubs ML. Allergy in 7000 twin pairs. Acta Allergol. 1971; 26:249–285.
Article5. Zielenski J, Tsui LC. Cystic fibrosis: genotypic and phenotypic variations. Annu Rev Genet. 1995; 29:777–807.
Article6. Van Eerdewegh P, Little RD, Dupuis J, Del Mastro RG, Falls K, Simon J, et al. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature. 2002; 418:426–430.
Article7. Allen M, Heinzmann A, Noguchi E, Abecasis G, Broxholme J, Ponting CP, et al. Positional cloning of a novel gene influencing asthma from chromosome 2q14. Nat Genet. 2003; 35:258–263.
Article8. Zhang Y, Leaves NI, Anderson GG, Ponting CP, Broxholme J, Holt R, et al. Positional cloning of a quantitative trait locus on chromosome 13q14 that influences immunoglobulin E levels and asthma. Nat Genet. 2003; 34:181–186.
Article9. Laitinen T, Polvi A, Rydman P, Vendelin J, Pulkkinen V, Salmikangas P, et al. Characterization of a common susceptibility locus for asthma-related traits. Science. 2004; 304:300–304.
Article10. Oguma T, Palmer LJ, Birben E, Sonna LA, Asano K, Lilly CM. Role of prostanoid DP receptor variants in susceptibility to asthma. N Engl J Med. 2004; 351:1752–1763.
Article11. Risch NJ. Searching for genetic determinants in the new millennium. Nature. 2000; 405:847–856.
Article12. Hardy J, Singleton A. Genomewide association studies and human disease. N Engl J Med. 2009; 360:1759–1768.
Article13. Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007; 448:470–473.
Article14. Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010; 363:1211–1221.
Article15. Sheehan NA, Didelez V, Burton PR, Tobin MD. Mendelian randomisation and causal inference in observational epidemiology. PLoS Med. 2008; 5:e177.
Article16. Levin AM, Mathias RA, Huang L, Roth LA, Daley D, Myers RA, et al. A meta-analysis of genome-wide association studies for serum total IgE in diverse study populations. J Allergy Clin Immunol. 2013; 131:1176–1184.
Article17. Kim JH, Cheong HS, Park JS, Jang AS, Uh ST, Kim YH, et al. A genome-wide association study of total serum and mite-specific IgEs in asthma patients. PLoS One. 2013; 8:e71958.
Article18. Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009; 461:747–753.
Article19. 1000 Genomes Project Consortium. Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 2010; 467:1061–1073.
Article20. Raska P, Zhu X. Rare variant density across the genome and across populations. BMC Proc. 2011; 5:Suppl 9. S39.
Article21. Ortega VE, Hawkins GA, Moore WC, Hastie AT, Ampleford EJ, Busse WW, et al. Effect of rare variants in ADRB2 on risk of severe exacerbations and symptom control during longacting β agonist treatment in a multiethnic asthma population: a genetic study. Lancet Respir Med. 2014; 2:204–213.
Article22. Halapi E, Gudbjartsson DF, Jonsdottir GM, Bjornsdottir US, Thorleifsson G, Helgadottir H, et al. A sequence variant on 17q21 is associated with age at onset and severity of asthma. Eur J Hum Genet. 2010; 18:902–908.
Article23. Green RH, Brightling CE, Bradding P. The reclassification of asthma based on subphenotypes. Curr Opin Allergy Clin Immunol. 2007; 7:43–50.
Article24. Moore WC, Meyers DA, Wenzel SE, Teague WG, Li H, Li X, et al. Identification of asthma phenotypes using cluster analysis in the Severe Asthma Research Program. Am J Respir Crit Care Med. 2010; 181:315–323.
Article25. Jang AS, Kwon HS, Cho YS, Bae YJ, Kim TB, Park JS, et al. Identification of subtypes of refractory asthma in Korean patients by cluster analysis. Lung. 2013; 191:87–93.
Article26. Ho SM. Environmental epigenetics of asthma: an update. J Allergy Clin Immunol. 2010; 126:453–465.
Article27. Thomsen SF, van der Sluis S, Kyvik KO, Skytthe A, Backer V. Estimates of asthma heritability in a large twin sample. Clin Exp Allergy. 2010; 40:1054–1061.
Article28. Haldane JBS. Heredity and politics. London: George Allen & Unwin Ltd Pblishe;1938.29. Huiyan W, Yuhe G, Juan W, Junyan Z, Shan W, Xiaojun Z, et al. The Importance of Allergen Avoidance in High Risk Infants and Sensitized Patients: A Meta-analysis Study. Allergy Asthma Immunol Res. 2014; 6:525–534.
Article30. Peden D, Reed CE. Environmental and occupational allergies. J Allergy Clin Immunol. 2010; 125:2 Suppl 2. S150–S160.
Article31. von Mutius E. Gene-environment interactions in asthma. J Allergy Clin Immunol. 2009; 123:3–11.
Article32. Park HJ, Lim HS, Park KH, Lee JH, Park JW, Hong CS. Changes in allergen sensitization over the last 30 years in Korea respiratory allergic patients: a single-center. Allergy Asthma Immunol Res. 2014; 6:434–443.
Article33. Lee JH, Lee HS, Park MR, Lee SW, Kim EH, Cho JB, et al. Relationship between indoor air pollutant levels and residential environment in children with atopic dermatitis. Allergy Asthma Immunol Res. 2014; 6:517–524.
Article34. Jeong I, Kim I, Park HJ, Roh J, Park JW, Lee JH. Allergic diseases and multiple chemical sensitivity in Korean adults. Allergy Asthma Immunol Res. 2014; 6:409–414.
Article35. Hur GY, Ye YM, Koh DH, Kim SH, Park HS. IL-4 Receptor α Polymorphisms May Be a Susceptible Factor for Work-Related Respiratory Symptoms in Bakery Workers. Allergy Asthma Immunol Res. 2013; 5:371–376.
Article36. Yang SN, Hsieh CC, Kuo HF, Lee MS, Huang MY, Kuo CH, et al. The effects of environmental toxins on allergic inflammation. Allergy Asthma Immunol Res. 2014; 6:478–484.
Article37. Kim BJ, Lee SY, Kim HB, Lee E, Hong SJ. Environmental changes, microbiota, and allergic diseases. Allergy Asthma Immunol Res. 2014; 6:389–400.
Article38. Yang SI, Lee E, Jung YH, Kim HY, Seo JH, Kwon JW, et al. Effect of antibiotic use and mold exposure in infancy on allergic rhinitis in susceptible adolescents. Ann Allergy Asthma Immunol. 2014; 113:160–165.e1.
Article39. Kim SH, Sutherland ER, Gelfand EW. Is there a link between obesity and asthma? Allergy Asthma Immunol Res. 2014; 6:189–195.
Article40. Lee SY, Kang MJ, Kwon JW, Park KS, Hong SJ. Breastfeeding Might Have Protective Effects on Atopy in Children With the CD14C-159T CT/CC Genotype. Allergy Asthma Immunol Res. 2013; 5:239–241.
Article41. Seo S, Kim D, Paul C, Yoo Y, Choung JT. Exploring Household-level Risk Factors for Self-reported Prevalence of Allergic Diseases Among Low-income Households in Seoul, Korea. Allergy Asthma Immunol Res. 2014; 6:421–427.
Article42. Zhang G, Khoo SK, Mäkelä MJ, Candelaria P, Hayden CM, von Hertzen L, et al. Maternal Genetic Variants of IL4/IL13 Pathway Genes on IgE With "Western or Eastern Environments/Lifestyles". Allergy Asthma Immunol Res. 2014; 6:350–356.
Article43. Vercelli D. Learning from discrepancies: CD14 polymorphisms, atopy and the endotoxin switch. Clin Exp Allergy. 2003; 33:153–155.
Article44. Martinez FD. CD14, endotoxin, and asthma risk: actions and interactions. Proc Am Thorac Soc. 2007; 4:221–225.
Article45. Aderem A, Ulevitch RJ. Toll-like receptors in the induction of the innate immune response. Nature. 2000; 406:782–787.
Article46. Howell WM, Holgate ST. HLA genetics and allergic disease. Thorax. 1995; 50:815–818.
Article47. Kim SH, Cho BY, Park CS, Shin ES, Cho EY, Yang EM, et al. Alpha-T-catenin (CTNNA3) gene was identified as a risk variant for toluene diisocyanate-induced asthma by genome-wide association analysis. Clin Exp Allergy. 2009; 39:203–212.
Article48. Park SM, Park JS, Park HS, Park CS. Unraveling the genetic basis of aspirin hypersensitivity in asthma beyond arachidonate pathways. Allergy Asthma Immunol Res. 2013; 5:258–276.
Article49. Park BL, Kim TH, Kim JH, Bae JS, Pasaje CF, Cheong HS, et al. Genome-wide association study of aspirin-exacerbated respiratory disease in a Korean population. Hum Genet. 2013; 132:313–321.
Article50. Bouzigon E, Corda E, Aschard H, Dizier MH, Boland A, Bousquet J, et al. Effect of 17q21 variants and smoking exposure in early-onset asthma. N Engl J Med. 2008; 359:1985–1994.
Article51. Flory JH, Sleiman PM, Christie JD, Annaiah K, Bradfield J, Kim CE, et al. 17q12-21 variants interact with smoke exposure as a risk factor for pediatric asthma but are equally associated with early-onset versus late-onset asthma in North Americans of European ancestry. J Allergy Clin Immunol. 2009; 124:605–607.
Article52. van der Valk RJ, Duijts L, Kerkhof M, Willemsen SP, Hofman A, Moll HA, et al. Interaction of a 17q12 variant with both fetal and infant smoke exposure in the development of childhood asthma-like symptoms. Allergy. 2012; 67:767–774.
Article53. Ege MJ, Strachan DP, Cookson WO, Moffatt MF, Gut I, Lathrop M, et al. Gene-environment interaction for childhood asthma and exposure to farming in Central Europe. J Allergy Clin Immunol. 2011; 127:138–144. 144.e1-4.
Article54. Hamza TH, Chen H, Hill-Burns EM, Rhodes SL, Montimurro J, Kay DM, et al. Genome-wide gene-environment study identifies glutamate receptor gene GRIN2A as a Parkinson's disease modifier gene via interaction with coffee. PLoS Genet. 2011; 7:e1002237.
Article55. Wild CP. The exposome: from concept to utility. Int J Epidemiol. 2012; 41:24–32.
Article56. Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat Med. 2012; 18:716–725.
Article57. Bookman EB, McAllister K, Gillanders E, Wanke K, Balshaw D, Rutter J, et al. Gene-environment interplay in common complex diseases: forging an integrative model-recommendations from an NIH workshop. Genet Epidemiol. 2011; 35:217–225.
Article58. Thomas D. Gene--environment-wide association studies: emerging approaches. Nat Rev Genet. 2010; 11:259–272.
Article59. Han MK, Huang YJ, Lipuma JJ, Boushey HA, Boucher RC, Cookson WO, et al. Significance of the microbiome in obstructive lung disease. Thorax. 2012; 67:456–463.
Article60. Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007; 128:635–638.
Article61. Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006; 31:89–97.
Article62. Sato S, Yagi S, Arai Y, Hirabayashi K, Hattori N, Iwatani M, et al. Genome-wide DNA methylation profile of tissue-dependent and differentially methylated regions (T-DMRs) residing in mouse pluripotent stem cells. Genes Cells. 2010; 15:607–618.
Article63. Cheong HS, Park SM, Kim MO, Park JS, Lee JY, Byun JY, et al. Genome-wide methylation profile of nasal polyps: relation to aspirin hypersensitivity in asthmatics. Allergy. 2011; 66:637–644.
Article64. Kim YJ, Park SW, Kim TH, Park JS, Cheong HS, Shin HD, et al. Genome-wide methylation profiling of the bronchial mucosa of asthmatics: relationship to atopy. BMC Med Genet. 2013; 14:39.
Article65. Gudbjartsson DF, Bjornsdottir US, Halapi E, Helgadottir A, Sulem P, Jonsdottir GM, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009; 41:342–347.66. Himes BE, Hunninghake GM, Baurley JW, Rafaels NM, Sleiman P, Strachan DP, et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Genet. 2009; 84:581–593.
Article67. Mashimo T, Hadjebi O, Amair-Pinedo F, Tsurumi T, Langa F, Serikawa T, et al. Progressive Purkinje cell degeneration in tambaleante mutant mice is a consequence of a missense mutation in HERC1 E3 ubiquitin ligase. PLoS Genet. 2009; 5:e1000784.
Article68. Sleiman PM, Flory J, Imielinski M, Bradfield JP, Annaiah K, Willis-Owen SA, et al. Variants of DENND1B associated with asthma in children. N Engl J Med. 2010; 362:36–44.
Article69. Li X, Howard TD, Zheng SL, Haselkorn T, Peters SP, Meyers DA, et al. Genome-wide association study of asthma identifies RAD50-IL13 and HLA-DR/DQ regions. J Allergy Clin Immunol. 2010; 125:328–335.e11.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Asthma and epigenetics
- Gene-Environment Interactions Should be Considered in Future Studies to Understand the Association Between Prenatal Folate Supplementation and Asthma Development
- Gene - Gene Interactions Among MCP Genes Polymorphisms in Asthma
- HOTAIR Long Non-coding RNA: Characterizing the Locus Features by the In Silico Approaches
- Occupational Asthma: Etiologies and Risk Factors