J Periodontal Implant Sci.
2015 Apr;45(2):69-75. 10.5051/jpis.2015.45.2.69.
Multiple transcripts of anoctamin genes expressed in the mouse submandibular salivary gland
- Affiliations
-
- 1Program in Neurobiology, Seoul National University School of Dentistry and Dental Research Institute, Seoul, Korea. frankyu@snu.ac.kr
- 2Department of Conservative Dentistry, Seoul National University School of Dentistry, Seoul, Korea.
- 3Department of Oral Biochemistry, Seoul National University School of Dentistry, Seoul, Korea.
- 4Laboratory of Veterinary Biochemistry and Molecular Biology, Chungbuk National University College of Veterinary Medicine, Cheongju, Korea.
- KMID: 2329658
- DOI: http://doi.org/10.5051/jpis.2015.45.2.69
Abstract
- PURPOSE
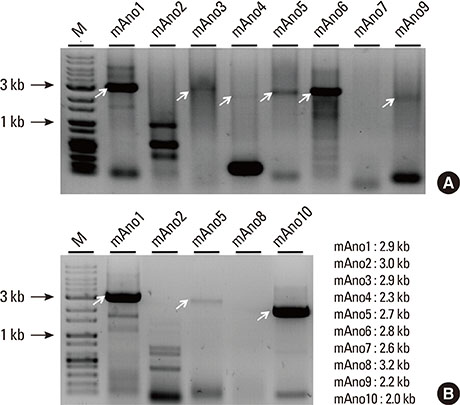
Salivary fluid formation is primarily driven by Ca2+-activated, apical efflux of chloride into the lumen of the salivary acinus. The anoctamin1 protein is an anion channel with properties resembling the endogenous calcium-activated chloride channels. In order to better understand the role of anoctamin proteins in salivary exocrine secretion, the expression of the ten members of the anoctamin gene family in the mouse submandibular gland was studied.
METHODS
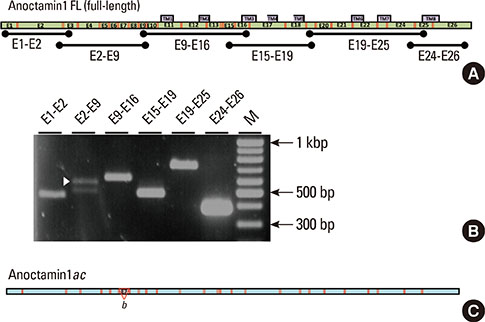
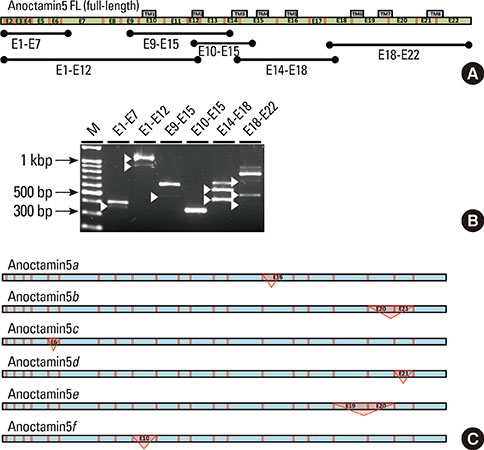
Total RNA extracted from mouse submandibular salivary glands was reverse transcribed using primer pairs to amplify the full-length coding regions of each anoctamin gene and was subcloned into plasmid vectors for DNA sequencing. Alternative splice variants were also screened by polymerase chain reaction using primer pairs that amplified six overlapping regions of the complementary DNA of each anoctamin gene, spanning multiple exons.
RESULTS
Multiple anoctamin transcripts were found in the mouse submandibular salivary gland, including full-length transcripts of anoctamin1, anoctamin3, anoctamin4, anoctamin5, anoctamin6, anoctamin9, and anoctamin10. Exon-skipping splicing in the N-terminal exons of the anoctamins1, anoctamin5, and anoctamin6 genes resulted in multiple alternative splice variants. No expression of anoctamin2, anoctamin7, or anoctamin8 was found.
CONCLUSIONS
The predominant anoctamin transcript expressed in the mouse submandibular gland is anoctamin1ac. The chloride channel protein produced by anoctamin1ac is likely responsible for the Ca2+-activated chloride efflux, which is the rate-limiting step in salivary exocrine secretion.
MeSH Terms
Figure
Reference
-
1. Smith PM. Mechanisms of salivary secretion. In : Edgar M, Dawes C, O'Mullane D, editors. Saliva and oral health: an essential overview for the health professional. 4th ed. Duns Tew: Stephen Hancocks Ltd;2012. p. 17–36.2. Yang YD, Cho H, Koo JY, Tak MH, Cho Y, Shim WS, et al. TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature. 2008; 455:1210–1215.
Article3. Schroeder BC, Cheng T, Jan YN, Jan LY. Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell. 2008; 134:1019–1029.
Article4. Caputo A, Caci E, Ferrera L, Pedemonte N, Barsanti C, Sondo E, et al. TMEM16A, a membrane protein associated with calcium-dependent chloride channel activity. Science. 2008; 322:590–594.
Article5. Duran C, Hartzell HC. Physiological roles and diseases of Tmem16/Anoctamin proteins: are they all chloride channels? Acta Pharmacol Sin. 2011; 32:685–692.
Article6. Tsutsumi S, Kamata N, Vokes TJ, Maruoka Y, Nakakuki K, Enomoto S, et al. The novel gene encoding a putative transmembrane protein is mutated in gnathodiaphyseal dysplasia (GDD). Am J Hum Genet. 2004; 74:1255–1261.
Article7. Bolduc V, Marlow G, Boycott KM, Saleki K, Inoue H, Kroon J, et al. Recessive mutations in the putative calcium-activated chloride channel Anoctamin 5 cause proximal LGMD2L and distal MMD3 muscular dystrophies. Am J Hum Genet. 2010; 86:213–221.
Article8. Suzuki J, Umeda M, Sims PJ, Nagata S. Calcium-dependent phospholipid scrambling by TMEM16F. Nature. 2010; 468:834–838.
Article9. Vermeer S, Hoischen A, Meijer RP, Gilissen C, Neveling K, Wieskamp N, et al. Targeted next-generation sequencing of a 12.5 Mb homozygous region reveals ANO10 mutations in patients with autosomal-recessive cerebellar ataxia. Am J Hum Genet. 2010; 87:813–819.
Article10. Ferrera L, Caputo A, Ubby I, Bussani E, Zegarra-Moran O, Ravazzolo R, et al. Regulation of TMEM16A chloride channel properties by alternative splicing. J Biol Chem. 2009; 284:33360–33368.
Article11. Sondo E, Scudieri P, Tomati V, Caci E, Mazzone A, Farrugia G, et al. Non-canonical translation start sites in the TMEM16A chloride channel. Biochim Biophys Acta. 2014; 1838:89–97.
Article12. Romanenko VG, Catalán MA, Brown DA, Putzier I, Hartzell HC, Marmorstein AD, et al. Tmem16A encodes the Ca2+-activated Cl- channel in mouse submandibular salivary gland acinar cells. J Biol Chem. 2010; 285:12990–13001.
Article13. Schreiber R, Uliyakina I, Kongsuphol P, Warth R, Mirza M, Martins JR, et al. Expression and function of epithelial anoctamins. J Biol Chem. 2010; 285:7838–7845.
Article14. Shimizu T, Iehara T, Sato K, Fujii T, Sakai H, Okada Y. TMEM16F is a component of a Ca2+-activated Cl- channel but not a volume-sensitive outwardly rectifying Cl- channel. Am J Physiol Cell Physiol. 2013; 304:C748–C759.15. Yang H, Jin T, Cheng T, Jan YN, Jan LY. Scan: a novel small-conductance Ca2+-activated non-selective cation channel encoded by TMEM16F. Biophys J. 2011; 100:259a.
Article16. Yang H, Kim A, David T, Palmer D, Jin T, Tien J, et al. TMEM16F forms a Ca2+-activated cation channel required for lipid scrambling in platelets during blood coagulation. Cell. 2012; 151:111–122.
Article17. Huang F, Wang X, Ostertag EM, Nuwal T, Huang B, Jan YN, et al. TMEM16C facilitates Na(+)-activated K+ currents in rat sensory neurons and regulates pain processing. Nat Neurosci. 2013; 16:1284–1290.
Article18. Tian Y, Kongsuphol P, Hug M, Ousingsawat J, Witzgall R, Schreiber R, et al. Calmodulin-dependent activation of the epithelial calcium-dependent chloride channel TMEM16A. FASEB J. 2011; 25:1058–1068.
Article19. Jung J, Nam JH, Park HW, Oh U, Yoon JH, Lee MG. Dynamic modulation of ANO1/TMEM16A HCO3(-) permeability by Ca2+/calmodulin. Proc Natl Acad Sci U S A. 2013; 110:360–365.20. Duran C, Qu Z, Osunkoya AO, Cui Y, Hartzell HC. ANOs 3-7 in the anoctamin/Tmem16 Cl- channel family are intracellular proteins. Am J Physiol Cell Physiol. 2012; 302:C482–C493.
Article21. Mahjneh I, Jaiswal J, Lamminen A, Somer M, Marlow G, Kiuru-Enari S, et al. A new distal myopathy with mutation in anoctamin 5. Neuromuscul Disord. 2010; 20:791–795.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A Case of Multiple Sialoliths in Sublingual Gland Misdiagnosed as Sialoliths in Submandibular Gland
- A Case of Removal of Multiple Submandibular Gland and Duct Stones by Cervical Incision Approach
- Recurrent Sialolithiasis on Remnant Wharton's Duct Following Submandibular Gland Resection
- Salivary Duct Carcinoma: 2 Case Reports
- A giant sialolith in a wharton's duct: A case report