Nutr Res Pract.
2011 Aug;5(4):288-293.
Radish (Raphanus sativus L. leaf) ethanol extract inhibits protein and mRNA expression of ErbB2 and ErbB3 in MDA-MB-231 human breast cancer cells
- Affiliations
-
- 1Department of Food Science and Nutrition, Dankook University, Gyeonggi 448-701, Korea.
- 2Department of Food and Nutrition, Kookmin University, 861-1 Chongneung-dong, Sungbuk-gu, Seoul 136-702, Korea. cmoon@kookmin.ac.kr
- 3Functional Food and Nutrition Division, Rural National Academy of Agricultural Science, Suwon 441-707, Korea.
Abstract
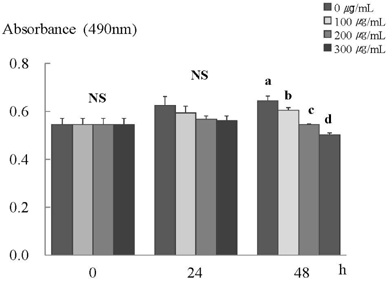
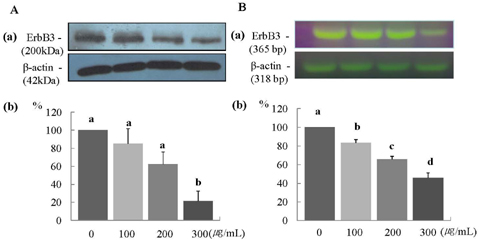
- In this study, we investigated the effects of the ethanol extract of aerial parts of Raphanus sativus L. (ERL) on breast cancer cell proliferation and gene expression associated with cell proliferation and apoptosis in MDA-MB-231 human breast cancer cells. The MDA-MB-231 cells were cultured in the presence or absence of various concentrations (100, 200, or 300 microg/mL) of ERL. ERL significantly decreased cell proliferation after 48 h of incubation (P < 0.05). The protein and mRNA expression of ErbB2 were decreased significantly in a dose-dependent manner (P < 0.05). The protein expression of ErbB3 was decreased significantly at an ERL concentration of 300 microg/mL (P < 0.05), and mRNA expression of ErbB3 was decreased significantly in a dose-dependent manner (P < 0.05). The protein expression of Akt was decreased significantly at the ERL concentration of 200 microg/mL (P < 0.05), and the protein expression of pAkt was decreased significantly in a dose-dependent manner (P < 0.05). The mRNA expression of Akt was decreased significantly at the ERL concentration of 200 microg/mL ERL (P < 0.05). The protein and mRNA expression of Bax were increased significantly at ERL concentrations of 200 microg/mL or higher (P < 0.05). The protein expression of Bcl2 was increased significantly at ERL concentrations of 100 microg/mL or higher (P < 0.05), and mRNA expression of Bcl2 was increased significantly at an ERL concentration of 300 microg/mL (P < 0.05). In conclusion, we suggest that Raphanus sativus, L. inhibits cell proliferation via the ErbB-Akt pathway in MDA-MB-231 cells.
Keyword
MeSH Terms
Figure
Reference
-
1. Ferlay J, Shin H, Bray F, Forman D, Mathers C, Parkin D. GLOBOCAN 2008, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 10. 2010. Lyon, France: International Agency for Research on Cancer.2. Annual report of cancer statistics in Korea in 2008. National Cancer Center [Internet]. cited 2011 March 2. Available from: http://ncc.re.kr/common/downloadByNTC.jsp?attnum=232&code=999_101.3. Korea Health Statistics 2009, Korea National Health and Nutrition Examination Survey (KNHANES IV-3). Korea Institute of Health and Social Affairs, Ministry of Health and Welfare Affairs [Internet]. cited 2011 May 27. Available from: http://knhanes.cdc.go.kr/.4. Kim BR, Park JH, Kim SH, Cho KJ, Chang MJ. Antihypertensive properties of dried radish leaves powder in spontaneously hypertensive rats. Korean J Nutr. 2010. 43:561–569.
Article5. Beevi SS, Narasu ML, Gowda BB. Polyphenolics profile, antioxidant and radical scavenging activity of leaves and stem of Raphanus sativus L. Plant Foods Hum Nutr. 2010. 65:8–17.
Article6. Scalbert A, Williamson G. Dietary intake and bioavailability of polyphenols. J Nutr. 2000. 130:2073S–2085S.
Article7. Yang CS, Wang ZY. Tea and cancer. J Natl Cancer Inst. 1993. 85:1038–1049.
Article8. Seeram NP, Adams LS, Zhang Y, Lee R, Sand D, Scheuller HS, Heber D. Blackberry, black raspberry, blueberry, cranberry, red raspberry, and strawberry extracts inhibit growth and stimulate apoptosis of human cancer cells in vitro. J Agric Food Chem. 2006. 54:9329–9339.
Article9. Pianetti S, Guo S, Kavanagh KT, Sonenshein GE. Green tea polyphenol epigallocatechin-3 gallate inhibits Her-2/neu signaling, proliferation, and transformed phenotype of breast cancer cells. Cancer Res. 2002. 62:652–655.10. Kameoka S, Leavitt P, Chang C, Kuo SM. Expression of antioxidant proteins in human intestinal Caco-2 cells treated with dietary flavonoids. Cancer Lett. 1999. 146:161–167.
Article11. Seshadri S, Nambiar VS. Simopoulos AP, Gopalan C, editors. Kanjero (Digera arvensis) and drumstick leaves (Moringa oleifera): nutrient profile and potential for human consumption. Plants in Human Health and Nutrition Policy. 2003. 41–59.
Article12. Papi A, Orlandi M, Bartolini G, Barillari J, Iori R, Paolini M, Ferroni F, Grazia Fumo M, Pedulli GF, Valgimigli L. Cytotoxic and antioxidant activity of 4-methylthio-3-butenyl isothiocyanate from Raphanus sativus L. (Kaiware Daikon) sprouts. J Agric Food Chem. 2008. 56:875–883.
Article13. Callahan R. Genetic alterations in primary breast cancer. Breast Cancer Res Treat. 1989. 13:191–203.
Article14. Slamon DJ, Clark GM. Amplification of c-erbB-2 and aggressive human breast tumors? Science. 1988. 240:1795–1798.15. Takeuchi K, Ito F. EGF receptor in relation to tumor development: molecular basis of responsiveness of cancer cells to EGFR-targeting tyrosine kinase inhibitors. FEBS J. 2010. 277:316–326.
Article16. Burgess AW, Cho HS, Eigenbrot C, Ferguson KM, Garrett TP, Leahy DJ, Lemmon MA, Sliwkowski MX, Ward CW, Yokoyama S. An open-and-shut case? Recent insights into the activation of EGF/ErbB receptors. Mol Cell. 2003. 12:541–552.
Article17. Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006. 7:505–516.
Article18. Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989. 244:707–712.
Article19. Bold RJ, Termuhlen PM, McConkey DJ. Apoptosis, cancer and cancer therapy. Surg Oncol. 1997. 6:133–142.
Article20. Sen P, Mukherjee S, Ray D, Raha S. Involvement of the Akt/PKB signaling pathway with disease processes. Mol Cell Biochem. 2003. 253:241–246.21. Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987. 235:177–182.
Article22. Cooke T, Reeves J, Lannigan A, Stanton P. The value of the human epidermal growth factor receptor-2 (HER2) as a prognostic marker. Eur J Cancer. 2001. 37:3–10.
Article23. Lewis S, Locker A, Todd JH, Bell JA, Nicholson R, Elston CW, Blamey RW, Ellis IO. Expression of epidermal growth factor receptor in breast carcinoma. J Clin Pathol. 1990. 43:385–389.
Article24. Mendelsohn J, Baselga J. Status of epidermal growth factor receptor antagonists in the biology and treatment of cancer. J Clin Oncol. 2003. 21:2787–2799.
Article25. Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, Mardis E, Kupfer D, Wilson R, Kris M, Varmus H. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004. 101:13306–13311.
Article26. Janmaat ML, Kruyt FA, Rodriguez JA, Giaccone G. Response to epidermal growth factor receptor inhibitors in non-small cell lung cancer cells: limited antiproliferative effects and absence of apoptosis associated with persistent activity of extracellular signal-regulated kinase or Akt kinase pathways. Clin Cancer Res. 2003. 9:2316–2326.27. Yuan ZQ, Sun M, Feldman RI, Wang G, Ma X, Jiang C, Coppola D, Nicosia SV, Cheng JQ. Frequent activation of AKT2 and induction of apoptosis by inhibition of phosphoinositide-3-OH kinase/Akt pathway in human ovarian cancer. Oncogene. 2000. 19:2324–2330.
Article28. Bellacosa A, de Feo D, Godwin AK, Bell DW, Cheng JQ, Altomare DA, Wan M, Dubeau L, Scambia G, Masciullo V, Ferrandina G, Benedetti Panici P, Mancuso S, Neri G, Testa JR. Molecular alterations of the AKT2 oncogene in ovarian and breast carcinomas. Int J Cancer. 1995. 64:280–285.
Article29. Cardone MH, Roy N, Stennicke HR, Salvesen GS, Franke TF, Stanbridge E, Frisch S, Reed JC. Regulation of cell death protease caspase-9 by phosphorylation. Science. 1998. 282:1318–1321.
Article30. Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999. 96:857–868.
Article31. Tang ED, Nuñez G, Barr FG, Guan KL. Negative regulation of the forkhead transcription factor FKHR by Akt. J Biol Chem. 1999. 274:16741–16746.
Article32. Chun KH, Kosmeder JW 2nd, Sun S, Pezzuto JM, Lotan R, Hong WK, Lee HY. Effects of deguelin on the phosphatidylinositol 3-kinase/Akt pathway and apoptosis in premalignant human bronchial epithelial cells. J Natl Cancer Inst. 2003. 95:291–302.
Article33. Ow YP, Green DR, Hao Z, Mak TW. Cytochrome c: functions beyond respiration. Nat Rev Mol Cell Biol. 2008. 9:532–542.
Article34. Binder C, Marx D, Binder L, Schauer A, Hiddemann W. Expression of Bax in relation to Bcl-2 and other predictive parameters in breast cancer. Ann Oncol. 1996. 7:129–133.
Article35. Lee HS, Kim EJ, Kim SH. Chestnut extract induces apoptosis in AGS human gastric cancer cells. Nutr Res Pract. 2011. 5:185–191.
Article36. Sakakura C, Sweeney EA, Shirahama T, Igarashi Y, Hakomori S, Nakatani H, Tsujimoto H, Imanishi T, Ohgaki M, Ohyama T, Yamazaki J, Hagiwara A, Yamaguchi T, Sawai K, Takahashi T. Overexpression of bax sensitizes human breast cancer MCF-7 cells to radiation-induced apoptosis. Int J Cancer. 1996. 67:101–105.
Article37. Wolter KG, Hsu YT, Smith CL, Nechushtan A, Xi XG, Youle RJ. Movement of Bax from the cytosol to mitochondria during apoptosis. J Cell Biol. 1997. 139:1281–1292.
Article38. Tsuruta F, Masuyama N, Gotoh Y. The phosphatidylinositol 3-kinase (PI3K)-Akt pathway suppresses Bax translocation to mitochondria. J Biol Chem. 2002. 277:14040–14047.
Article39. Korsmeyer SJ, Wei MC, Saito M, Weiler S, Oh KJ, Schlesinger PH. Pro-apoptotic cascade activates BID, which oligomerizes BAK or BAX into pores that result in the release of cytochrome c. Cell Death Differ. 2000. 7:1166–1173.
Article40. Shimizu S, Narita M, Tsujimoto Y. Bcl-2 family proteins regulate the release of apoptogenic cytochrome c by the mitochondrial channel VDAC. Nature. 1999. 399:483–487.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Inorganic sulfur reduces cell proliferation by inhibiting of ErbB2 and ErbB3 protein and mRNA expression in MDA-MB-231 human breast cancer cells
- Effect of[6]-Gingerol on Inhibition of Cell Proliferation in MDA-MB-231 Human Breast Cancer Cells
- Loquat (Eriobotrya japonica) leaf extract inhibits the growth of MDA-MB-231 tumors in nude mouse xenografts and invasion of MDA-MB-231 cells
- Delphinidin inhibits cell proliferation and induces apoptosis in MDA-MB-231 human breast cancer cell lines
- Effects of alpha-lipoic acid on cell proliferation and apoptosis in MDA-MB-231 human breast cells