J Clin Neurol.
2012 Sep;8(3):230-234. 10.3988/jcn.2012.8.3.230.
Leber's Hereditary Optic Neuropathy with Olivocerebellar Degeneration due to G11778A and T3394C Mutations in the Mitochondrial DNA
- Affiliations
-
- 1Division of Neurology, Department of Brain and Neurosciences, Faculty of Medicine, Tottori University, Yonago, Japan. kazuhiro@med.tottori-u.ac.jp
- KMID: 2287596
- DOI: http://doi.org/10.3988/jcn.2012.8.3.230
Abstract
- BACKGROUND
Leber's hereditary optic neuropathy (LHON) is a mitochondrial disorder with optic nerve atrophy. Although there are no other associated neurological abnormalities in most cases of LHON, cases of "LHON plus" have been reported.
CASE REPORT
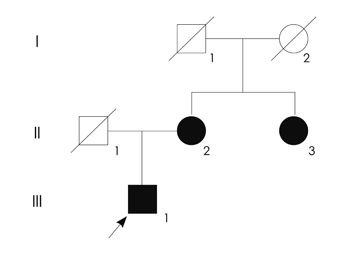
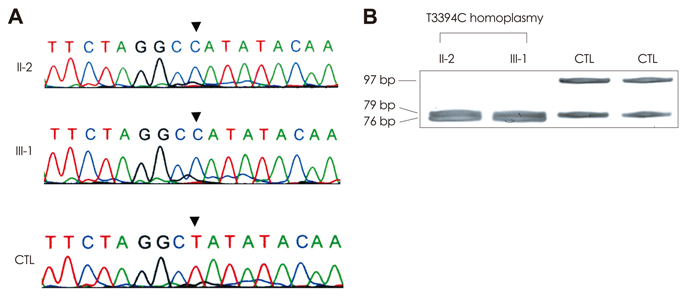
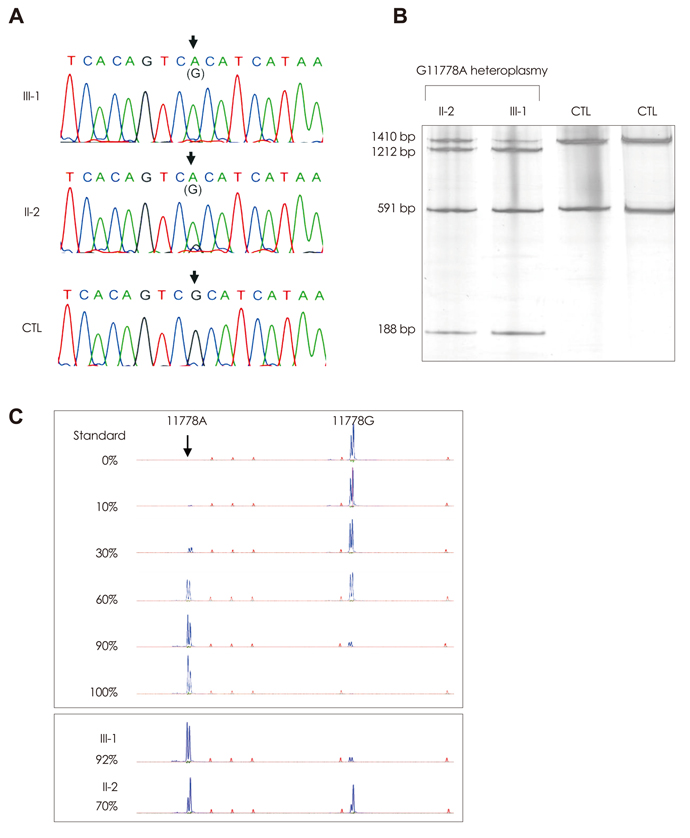
The proband was a 37-year-old man who had visual and gait disturbances that had first appeared at 10 years of age. He showed horizontal gaze palsy, gaze-evoked nystagmus, dysarthria, and cerebellar ataxia. Brain and orbit MRI disclosed atrophy of the optic nerve and cerebellum, and degenerative changes in the bilateral inferior olivary nucleus. Mutational analyses of mitochondrial DNA identified the coexistence of heteroplasmic G11778A and homoplasmic T3394C mutations.
CONCLUSIONS
These results suggest that the combination of G11778A and T3394C mutations leads to an atypical LHON phenotype.
Keyword
MeSH Terms
Figure
Reference
-
1. Carelli V, Achilli A, Valentino ML, Rengo C, Semino O, Pala M, et al. Haplogroup effects and recombination of mitochondrial DNA: novel clues from the analysis of Leber hereditary optic neuropathy pedigrees. Am J Hum Genet. 2006. 78:564–574.
Article2. Carelli V, Ross-Cisneros FN, Sadun AA. Mitochondrial dysfunction as a cause of optic neuropathies. Prog Retin Eye Res. 2004. 23:53–89.
Article3. Funakawa I, Kato H, Terao A, Ichihashi K, Kawashima S, Hayashi T, et al. Cerebellar ataxia in patients with Leber's hereditary optic neuropathy. J Neurol. 1995. 242:75–77.
Article4. Nikoskelainen EK, Marttila RJ, Huoponen K, Juvonen V, Lamminen T, Sonninen P, et al. Leber's "plus": neurological abnormalities in patients with Leber's hereditary optic neuropathy. J Neurol Neurosurg Psychiatry. 1995. 59:160–164.
Article5. La Morgia C, Achilli A, Iommarini L, Barboni P, Pala M, Olivieri A, et al. Rare mtDNA variants in Leber hereditary optic neuropathy families with recurrence of myoclonus. Neurology. 2008. 70:762–770.
Article6. Watanabe M, Mita S, Takita T, Goto Y, Uchino M, Imamura S. Leber's hereditary optic neuropathy with dystonia in a Japanese family. J Neurol Sci. 2006. 243:31–34.
Article7. Ruiz-Pesini E, Lott MT, Procaccio V, Poole JC, Brandon MC, Mishmar D, et al. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007. 35:D823–D828.
Article8. Man PY, Turnbull DM, Chinnery PF. Leber hereditary optic neuropathy. J Med Genet. 2002. 39:162–169.
Article9. Wallace DC, Singh G, Lott MT, Hodge JA, Schurr TG, Lezza AM, et al. Mitochondrial DNA mutation associated with Leber's hereditary optic neuropathy. Science. 1988. 242:1427–1430.
Article10. Carelli V, La Morgia C, Iommarini L, Carroccia R, Mattiazzi M, Sangiorgi S, et al. Mitochondrial optic neuropathies: how two genomes may kill the same cell type? Biosci Rep. 2007. 27:173–184.
Article11. Beretta S, Mattavelli L, Sala G, Tremolizzo L, Schapira AH, Martinuzzi A, et al. Leber hereditary optic neuropathy mtDNA mutations disrupt glutamate transport in cybrid cell lines. Brain. 2004. 127:2183–2192.
Article12. Chen H, Chan DC. Critical dependence of neurons on mitochondrial dynamics. Curr Opin Cell Biol. 2006. 18:453–459.
Article13. Murakami T, Mita S, Tokunaga M, Maeda H, Ueyama H, Kumamoto T, et al. Hereditary cerebellar ataxia with Leber's hereditary optic neuropathy mitochondrial DNA 11778 mutation. J Neurol Sci. 1996. 142:111–113.
Article14. Gropman A, Chen TJ, Perng CL, Krasnewich D, Chernoff E, Tifft C, et al. Variable clinical manifestation of homoplasmic G14459A mitochondrial DNA mutation. Am J Med Genet A. 2004. 124A:377–382.
Article15. Zhang AM, Jia X, Yao YG, Zhang Q. Co-occurrence of A1555G and G11778A in a Chinese family with high penetrance of Leber's hereditary optic neuropathy. Biochem Biophys Res Commun. 2008. 376:221–224.
Article16. Valentino ML, Barboni P, Ghelli A, Bucchi L, Rengo C, Achilli A, et al. The ND1 gene of complex I is a mutational hot spot for Leber's hereditary optic neuropathy. Ann Neurol. 2004. 56:631–641.
Article17. Liao WQ, Pang Y, Yu CA, Wen JY, Zhang YG, Li XH. Novel mutations of mitochondrial DNA associated with type 2 diabetes in Chinese Han population. Tohoku J Exp Med. 2008. 215:377–384.
Article18. Sciacco M, Prelle A, Fagiolari G, Bordoni A, Crimi M, Di Fonzo A, et al. A case of CPT deficiency, homoplasmic mtDNA mutation and ragged red fibers at muscle biopsy. J Neurol Sci. 2005. 239:21–24.
Article19. Wani AA, Ahanger SH, Bapat SA, Rangrez AY, Hingankar N, Suresh CG, et al. Analysis of mitochondrial DNA sequences in childhood encephalomyopathies reveals new disease-associated variants. PLoS One. 2007. 2:e942.
Article20. Arnestad M, Opdal SH, Vege A, Rognum TO. A mitochondrial DNA polymorphism associated with cardiac arrhythmia investigated in sudden infant death syndrome. Acta Paediatr. 2007. 96:206–210.
Article21. Wong LJ, Lin YH, Suwannarat P, Hsu CH, Kwon HY, Mackowiak S. Mitochondrial DNA mutations in a patient with sex reversal and clinical features consistent with Fraser syndrome. Clin Genet. 2005. 67:252–257.
Article22. Zhang M, Zhou X, Li C, Zhao F, Zhang J, Yuan M, et al. Mitochondrial haplogroup M9a specific variant ND1 T3394C may have a modifying role in the phenotypic expression of the LHON-associated ND4 G11778A mutation. Mol Genet Metab. 2010. 101:192–199.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Clinical Manifestations of Leber's Hereditary Optic Neuropathy with 11778 mtDNA Mutation
- A Mitochondrial Mutation in Leber's Hereditary Optic Neuropathy
- A Case of Leber's Hereditary Optic Nouropathy Showing 11778 Point Mutation of Mitochondrial DNA
- Ondine's Curse Consequent to Recurrent Respiratory Failure in a Man with Leber Hereditary Optic Neuropathy
- Optic Neuropathy in Koreans I. Leber's Hereditary Optic Neuropathy