Korean J Physiol Pharmacol.
2013 Aug;17(4):291-297. 10.4196/kjpp.2013.17.4.291.
Mutant p53-Notch1 Signaling Axis Is Involved in Curcumin-Induced Apoptosis of Breast Cancer Cells
- Affiliations
-
- 1Department of Dental Pharmacology, School of Dentistry, Yangsan Campus of Pusan National University, Yangsan 626-870, Korea. skbae@pusan.ac.kr
- 2Department of Oral Physiology, School of Dentistry, Yangsan Campus of Pusan National University, Yangsan 626-870, Korea.
- 3Korea Research Institute of Chemical Technology, Daejeon 305-600, Korea.
- 4College of Pharmacy, Seoul National University, Seoul 151-742, Korea.
- 5Severance Integrative Research Institute for Cerebral and Cardiovascular Diseases, Yonsei University Health System, Seoul 120-752, Korea.
- 6Medical Research Institute, Pusan National University, Busan 602-739, Korea.
- KMID: 2285463
- DOI: http://doi.org/10.4196/kjpp.2013.17.4.291
Abstract
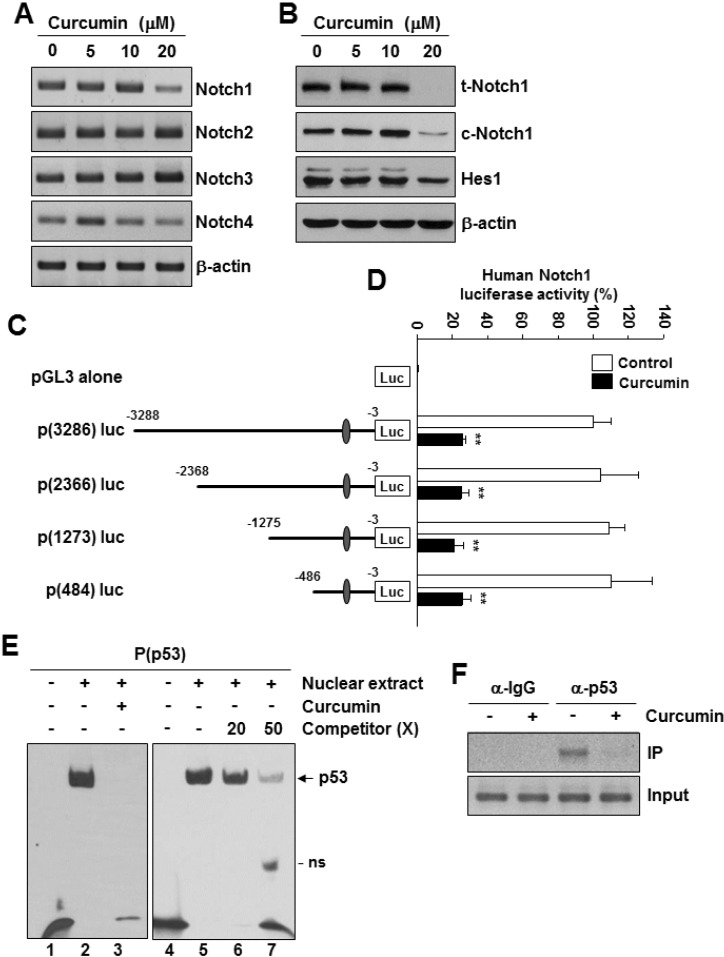
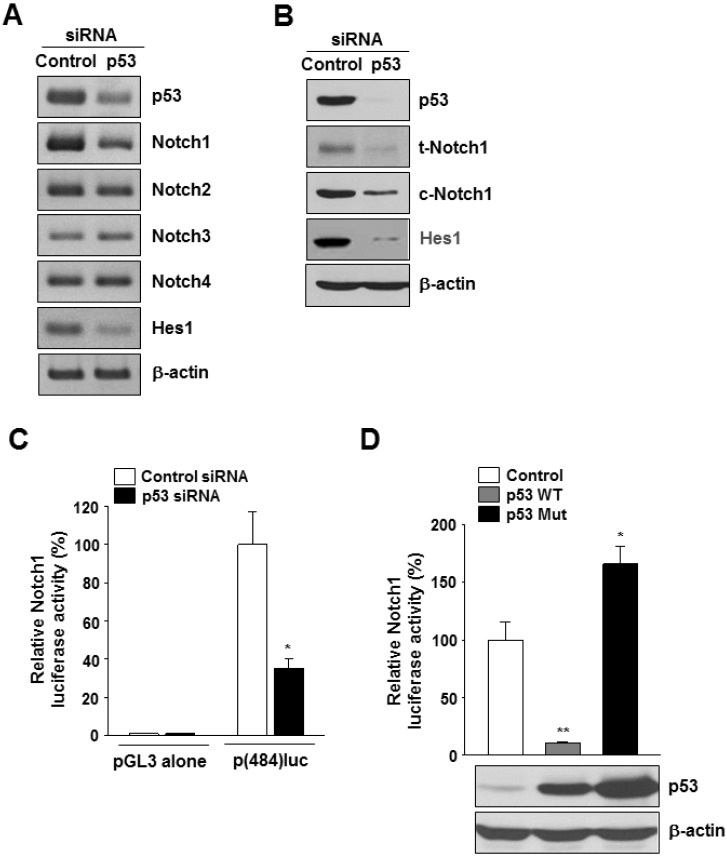
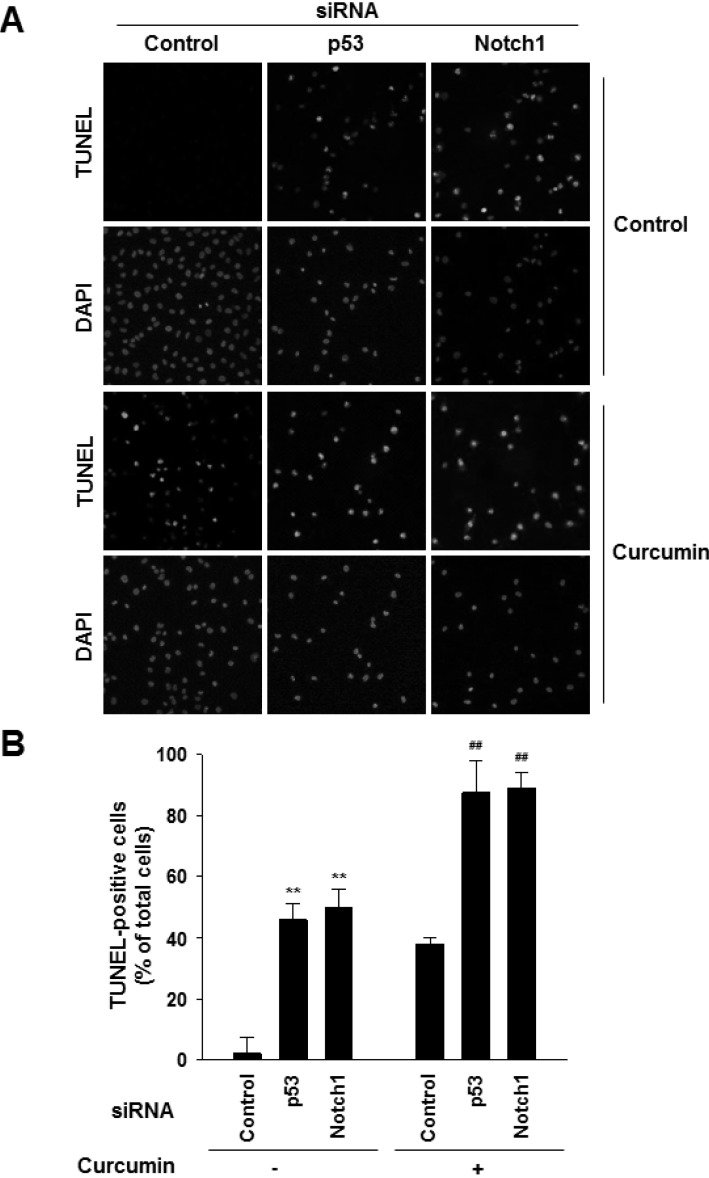
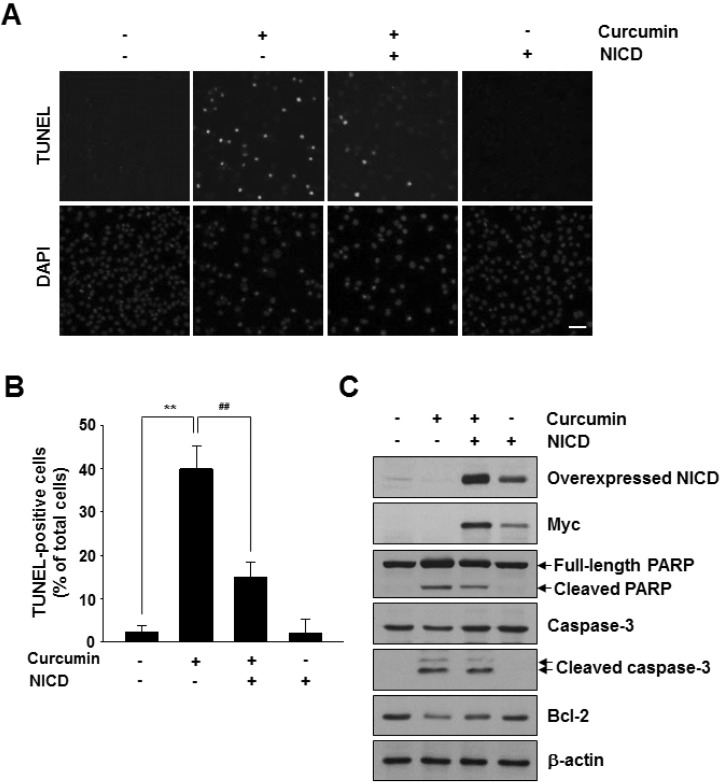
- Notch1 has been reported to be highly expressed in triple-negative and other subtypes of breast cancer. Mutant p53 (R280K) is overexpressed in MDA-MB-231 triple-negative human breast cancer cells. The present study aimed to determine whether the mutant p53 can be a potent transcriptional activator of the Notch1 in MDA-MB-231 cells, and explore the role of this mutant p53-Notch1 axis in curcumin-induced apoptosis. We found that curcumin treatment resulted in an induction of apoptosis in MDA-MB-231 cells, together with downregulation of Notch1 and its downstream target, Hes1. This reduction in Notch1 expression was determined to be due to the decreased activity of endogenous mutant p53. We confirmed the suppressive effect of curcumin on Notch1 transcription by performing a Notch1 promoter-driven reporter assay and identified a putative p53-binding site in the Notch1 promoter by EMSA and chromatin immunoprecipitation analysis. Overexpression of mutant p53 increased Notch1 promoter activity, whereas knockdown of mutant p53 by small interfering RNA suppressed Notch1 expression, leading to the induction of cellular apoptosis. Moreover, curcumin-induced apoptosis was further enhanced by the knockdown of Notch1 or mutant p53, but it was decreased by the overexpression of active Notch1. Taken together, our results demonstrate, for the first time, that Notch1 is a transcriptional target of mutant p53 in breast cancer cells and suggest that the targeting of mutant p53 and/or Notch1 may be combined with a chemotherapeutic strategy to improve the response of breast cancer cells to curcumin.
Keyword
MeSH Terms
Figure
Cited by 1 articles
-
Novel functional roles of caspase-related genes in the regulation of apoptosis and autophagy
Ju-Hyun Shin, Sang-Hyun Min
Korean J Physiol Pharmacol. 2016;20(6):573-580. doi: 10.4196/kjpp.2016.20.6.573.
Reference
-
1. Menon VP, Sudheer AR. Antioxidant and anti-inflammatory properties of curcumin. Adv Exp Med Biol. 2007; 595:105–125. PMID: 17569207.
Article2. Kunnumakkara AB, Anand P, Aggarwal BB. Curcumin inhibits proliferation, invasion, angiogenesis and metastasis of different cancers through interaction with multiple cell signaling proteins. Cancer Lett. 2008; 269:199–225. PMID: 18479807.
Article3. Kim SR, Park HJ, Bae YH, Ahn SC, Wee HJ, Yun I, Jang HO, Bae MK, Bae SK. Curcumin down-regulates visfatin expression and inhibits breast cancer cell invasion. Endocrinology. 2012; 153:554–563. PMID: 22186408.
Article4. Zhou H, Beevers CS, Huang S. The targets of curcumin. Curr Drug Targets. 2011; 12:332–347. PMID: 20955148.
Article5. Wang Z, Zhang Y, Banerjee S, Li Y, Sarkar FH. Notch-1 down-regulation by curcumin is associated with the inhibition of cell growth and the induction of apoptosis in pancreatic cancer cells. Cancer. 2006; 106:2503–2513. PMID: 16628653.6. Chen Y, Shu W, Chen W, Wu Q, Liu H, Cui G. Curcumin, both histone deacetylase and p300/CBP-specific inhibitor, represses the activity of nuclear factor kappa B and Notch 1 in Raji cells. Basic Clin Pharmacol Toxicol. 2007; 101:427–433. PMID: 17927689.
Article7. Liao S, Xia J, Chen Z, Zhang S, Ahmad A, Miele L, Sarkar FH, Wang Z. Inhibitory effect of curcumin on oral carcinoma CAL-27 cells via suppression of Notch-1 and NF-κB signaling pathways. J Cell Biochem. 2011; 112:1055–1065. PMID: 21308734.
Article8. Li Y, Zhang J, Ma D, Zhang L, Si M, Yin H, Li J. Curcumin inhibits proliferation and invasion of osteosarcoma cells through inactivation of Notch-1 signaling. FEBS J. 2012; 279:2247–2259. PMID: 22521131.
Article9. Penton AL, Leonard LD, Spinner NB. Notch signaling in human development and disease. Semin Cell Dev Biol. 2012; 23:450–457. PMID: 22306179.
Article10. Lobry C, Oh P, Aifantis I. Oncogenic and tumor suppressor functions of Notch in cancer: it's NOTCH what you think. J Exp Med. 2011; 208:1931–1935. PMID: 21948802.
Article11. Fassl A, Tagscherer KE, Richter J, Berriel Diaz M, Alcantara Llaguno SR, Campos B, Kopitz J, Herold-Mende C, Herzig S, Schmidt MH, Parada LF, Wiestler OD, Roth W. Notch1 signaling promotes survival of glioblastoma cells via EGFR-mediated induction of anti-apoptotic Mcl-1. Oncogene. 2012; 31:4698–4708. PMID: 22249262.
Article12. Simmons MJ, Serra R, Hermance N, Kelliher MA. NOTCH1 inhibition in vivo results in mammary tumor regression and reduced mammary tumorsphere-forming activity in vitro. Breast Cancer Res. 2012; 14:R126. [Epub ahead of print]. PMID: 22992387.
Article13. Speiser J, Foreman K, Drinka E, Godellas C, Perez C, Salhadar A, Erşahin Ç, Rajan P. Notch-1 and Notch-4 biomarker expression in triple-negative breast cancer. Int J Surg Pathol. 2012; 20:139–145. PMID: 22084425.
Article14. Selivanova G. p53: fighting cancer. Curr Cancer Drug Targets. 2004; 4:385–402. PMID: 15320716.
Article15. Horn HF, Vousden KH. Coping with stress: multiple ways to activate p53. Oncogene. 2007; 26:1306–1316. PMID: 17322916.
Article16. Molchadsky A, Rivlin N, Brosh R, Rotter V, Sarig R. p53 is balancing development, differentiation and de-differentiation to assure cancer prevention. Carcinogenesis. 2010; 31:1501–1508. PMID: 20504879.
Article17. Wijnhoven SW, Speksnijder EN, Liu X, Zwart E, vanOostrom CT, Beems RB, Hoogervorst EM, Schaap MM, Attardi LD, Jacks T, van Steeg H, Jonkers J, de Vries A. Dominant-negative but not gain-of-function effects of a p53.R270H mutation in mouse epithelium tissue after DNA damage. Cancer Res. 2007; 67:4648–4656. PMID: 17510390.18. Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009; 9:701–713. PMID: 19693097.
Article19. Walerych D, Napoli M, Collavin L, Del Sal G. The rebel angel: mutant p53 as the driving oncogene in breast cancer. Carcinogenesis. 2012; 33:2007–2017. PMID: 22822097.
Article20. Yugawa T, Handa K, Narisawa-Saito M, Ohno S, Fujita M, Kiyono T. Regulation of Notch1 gene expression by p53 in epithelial cells. Mol Cell Biol. 2007; 27:3732–3742. PMID: 17353266.
Article21. Lefort K, Mandinova A, Ostano P, Kolev V, Calpini V, Kolfschoten I, Devgan V, Lieb J, Raffoul W, Hohl D, Neel V, Garlick J, Chiorino G, Dotto GP. Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKalpha kinases. Genes Dev. 2007; 21:562–577. PMID: 17344417.22. Alimirah F, Panchanathan R, Davis FJ, Chen J, Choubey D. Restoration of p53 expression in human cancer cell lines upregulates the expression of Notch1: implications for cancer cell fate determination after genotoxic stress. Neoplasia. 2007; 9:427–434. PMID: 17534448.
Article23. Laws AM, Osborne BA. p53 regulates thymic Notch1 activation. Eur J Immunol. 2004; 34:726–734. PMID: 14991602.
Article24. Bocchetta M, Miele L, Pass HI, Carbone M. Notch-1 induction, a novel activity of SV40 required for growth of SV40-transformed human mesothelial cells. Oncogene. 2003; 22:81–89. PMID: 12527910.
Article25. Mehta SA, Christopherson KW, Bhat-Nakshatri P, Goulet RJ Jr, Broxmeyer HE, Kopelovich L, Nakshatri H. Negative regulation of chemokine receptor CXCR4 by tumor suppressor p53 in breast cancer cells: implications of p53 mutation or isoform expression on breast cancer cell invasion. Oncogene. 2007; 26:3329–3337. PMID: 17130833.
Article26. Bartek J, Iggo R, Gannon J, Lane DP. Genetic and immunochemical analysis of mutant p53 in human breast cancer cell lines. Oncogene. 1990; 5:893–899. PMID: 1694291.27. Gurtner A, Starace G, Norelli G, Piaggio G, Sacchi A, Bossi G. Mutant p53-induced up-regulation of mitogen-activated protein kinase kinase 3 contributes to gain of function. J Biol Chem. 2010; 285:14160–14169. PMID: 20223820.
Article28. Fontemaggi G, Dell'Orso S, Trisciuoglio D, Shay T, Melucci E, Fazi F, Terrenato I, Mottolese M, Muti P, Domany E, Del Bufalo D, Strano S, Blandino G. The execution of the transcriptional axis mutant p53, E2F1 and ID4 promotes tumor neo-angiogenesis. Nat Struct Mol Biol. 2009; 16:1086–1093. PMID: 19783986.
Article29. Yeudall WA, Vaughan CA, Miyazaki H, Ramamoorthy M, Choi MY, Chapman CG, Wang H, Black E, Bulysheva AA, Deb SP, Windle B, Deb S. Gain-of-function mutant p53 upregulates CXC chemokines and enhances cell migration. Carcinogenesis. 2012; 33:442–451. PMID: 22114072.
Article30. Petitjean A, Achatz MI, Borresen-Dale AL, Hainaut P, Olivier M. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene. 2007; 26:2157–2165. PMID: 17401424.
Article31. Lara-Medina F, Pérez-Sánchez V, Saavedra-Pérez D, Blake-Cerda M, Arce C, Motola-Kuba D, Villarreal-Garza C, González-Angulo AM, Bargalló E, Aguilar JL, Mohar A, Arrieta Ó. Triple-negative breast cancer in Hispanic patients: high prevalence, poor prognosis, and association with menopausal status, body mass index, and parity. Cancer. 2011; 117:3658–3669. PMID: 21387260.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- p53 signaling is involved in leptin-induced growth of hepatic and breast cancer cells
- Cellular Effect of Curcumin and Citral Combination on Breast Cancer Cells: Induction of Apoptosis and Cell Cycle Arrest
- The Role of bcl-2 and p53 in Tamoxifen-Induced Apoptosis of Human Breast Cancer Cell Lines
- Effect of Curcumin on Cancer Invasion and Matrix Metalloproteinase-9 Activity in MDA-MB-231 Human Breast Cancer Cell
- Translocation of Phospho-ser 15-p53 in Eugenol-induced Apoptosis of in vitro Cultured Cancer Cells