Infect Chemother.
2011 Oct;43(5):396-405. 10.3947/ic.2011.43.5.396.
Application of Infrequent-Restriction-Site Polymerase Reaction (IRS-PCR) to the Molecular Epidemiologic Analysis of Methicillin Resistant Staphylococcus aureus (MRSA)
- Affiliations
-
- 1Department of Biomedical Science, The Catholic University of Korea, Graduate School, Seoul, Korea.
- 2Department of Internal Medicine, College of Medicine, The Catholic University of Korea, Seoul, Korea. jhyoo@catholic.ac.kr
- 3Catholic Research Institutes of Medical Science, The Catholic University of Korea, Seoul, Korea.
- KMID: 2285012
- DOI: http://doi.org/10.3947/ic.2011.43.5.396
Abstract
- BACKGROUND
We investigated the usefulness of infrequent-restriction-site polymerase chain reaction (IRS-PCR) compared with pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST) on the molecular epidemiologic analysis of methicillin-resistant Staphylococcus aureus (MRSA).
MATERIALS AND METHODS
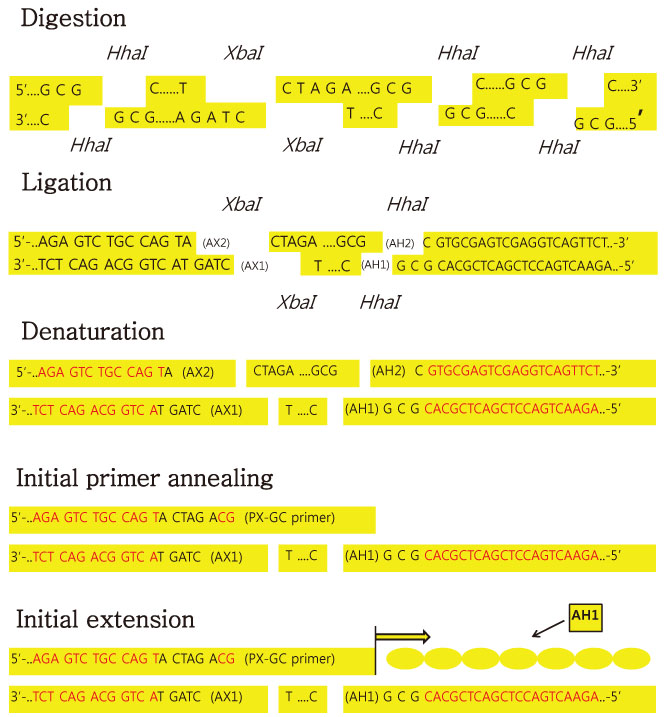
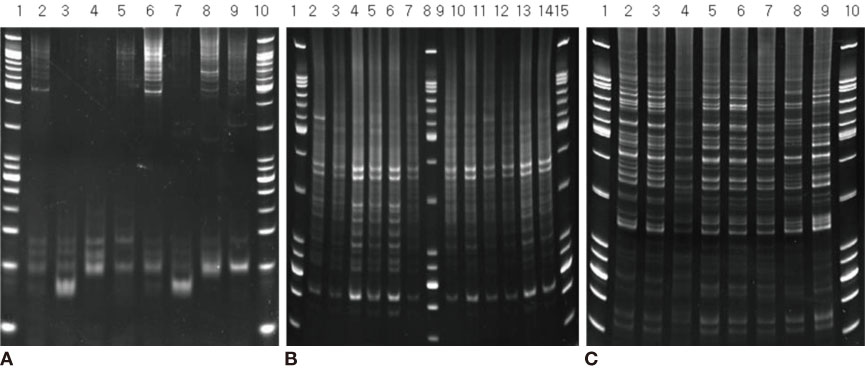
We used fifty clinical isolates of MRSA collected from 10 university hospitals located in Seoul. We performed three procedures on these isolates: PFGE using SmaI, IRS-PCR using XbaI-Hha I or EagI-Hha I, and MLST using seven house-keeping genes. We determined the clusters of molecular types by dendrogram using the unweighted pair group method with arithmetic mean (UPGMA) and Dice coefficients.
RESULTS
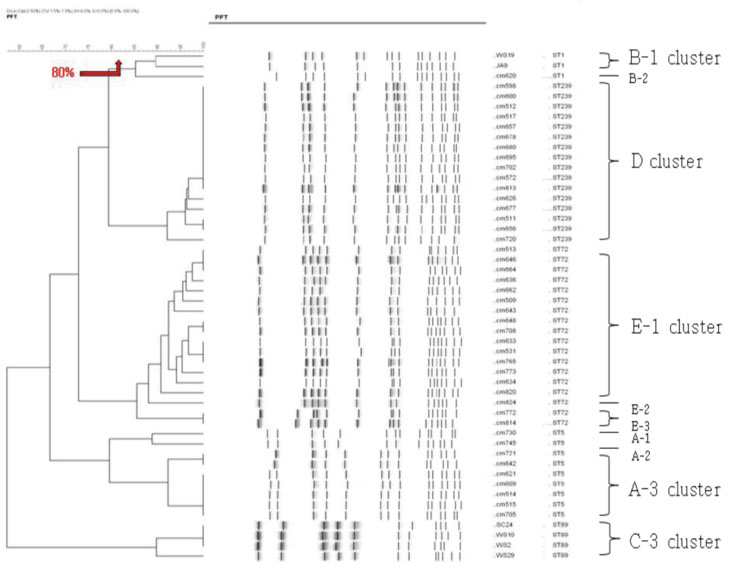
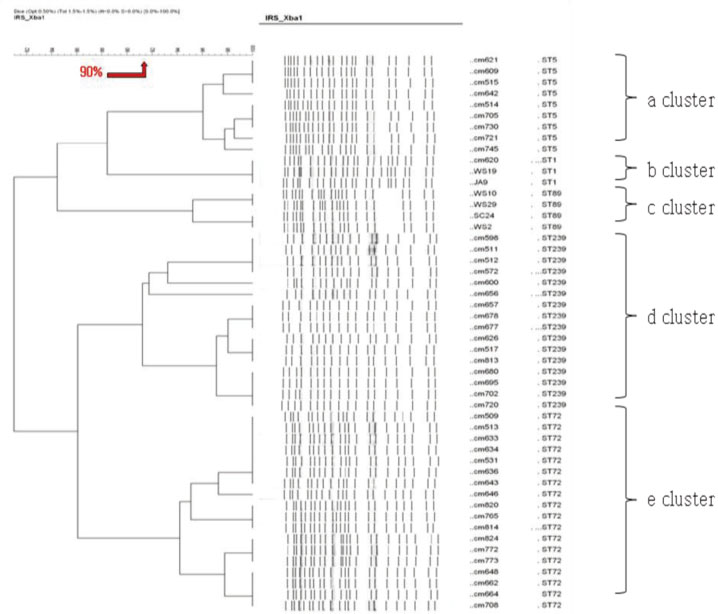
MLST analysis showed that isolates exhibited ST1, ST5, ST72, ST89, and ST239. In PFGE, the isolates clustered into 5 major groups with 80% similarity, which subsequently became classified into 18 subgroups with 95% similarity. In IRS-PCR using EagI-HhaI restriction enzymes, there was little resolution among the patterns of isolates. However, XbaI-HhaI IRS-PCR showed 5 groups with a 90% similarity. These groups were then classified into 9 subgroups with a 95% similarity. There were no significant differences among the isolates from different hospitals.
CONCLUSIONS
The XbaI-HhaI IRS-PCR method could be a useful tool in the molecular epidemiology of MRSA. Its resolution power was good enough to analyze isolates, because the patterns of IRS-PCR were closely correlated with those of MLST and showed diverse groups.
Keyword
MeSH Terms
Figure
Reference
-
1. Williams REO, Jevons MP, Shoote RA, Thom BT, Noble WC, Lidwell OM, White RG, Taylor GW. Isolation for the control of staphylococcal infection in surgical wards. Br Med J. 1962. 2:275–282.
Article2. Saravolatz LD, Markowitz N, Arking L, Pohlod D, Fisher E. Methicillin resistant Staphylococcus aureus. Epidemiologic observation during a community-acquired outbreak. Ann Intern Med. 1982. 96:11–16.3. Klein E, Smith DL, Laxminarayan R. Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999-2005. Emerg Infect Dis. 2007. 13:1840–1846.
Article4. Hussain FM, Boyle-Vavra S, Daum RS. Community-acquired methicillin-resistant Staphylococcus aureus colonization in healthy children attending an outpatient pediatric clinic. Pediatr Infect Dis J. 2001. 20:763–767.5. Suggs AH, Maranan MC, Boyle-Vavra S, Daum RS. Methicillin-resistant and borderline methicillin-resistant asymptomatic Staphylococcus aureus colonization in children without identifiable risk factors. Pediatr Infect Dis J. 1999. 18:410–414.
Article6. Miller LG, Perdreau-Remington F, Rieg G, Mehdi S, Perlroth J, Bayer AS, Tang AW, Phung TO, Spellberg B. Necrotizing fasciitis caused by community-associated methicillin-resistant Staphylococcus aureus in Los Angeles. N Engl J Med. 2005. 352:1445–1453.
Article7. Chong Y, Lee K. Present situation of antimicrobial resistance in Korea. J Infect Chemother. 2000. 6:189–195.
Article8. Kim JM, Park ES, Jeong JS, Kim KM, Kim JM, Oh HS, Yoon SW, Chang HS, Chang KH, Lee SI, Lee MS, Song JH, Kang MW, Park SC, Choe KW, Pai CH. Nosocomial Infection Surveillance Committee of the Korean Society for Nosocomial Infection Control. Multicenter surveillance study for nosocomial infections in major hospitals in Korea. Am J Infect Control. 2000. 28:454–458.
Article9. Kim HB, Sa CM, Yoo J, Kim BS, Yun OJ, Yoon HR, Lee YS. Antibiotic resistance patterns of Staphylococcus aureus isolated from the patients admitted to non-teriary hospitals. Korean J Infect Dis. 2000. 32:259–263.10. Kim SM, Lee DC, Park SD, Kim BS, Kim JK, Choi MR, Park SY, Hwang SM, Shin NY, Shim ES, Kwon PS, Kwon DY, Hur SH, Kim HJ, Lim HB, Chong Y. Genotype, coagulase type and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus isolated from dermatology patients and healthy individuals in Korea. J Bacteriol Virol. 2009. 39:307–316.
Article11. McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol. 2003. 41:5113–5120.
Article12. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000. 38:1008–1015.
Article13. Mazurek GH, Reddy V, Maston BJ, Haas WH, Crawford JT. DNA fingerprinting by infrequent-restriction-site amplification. J Clin Microbiol. 1996. 34:2386–2390.
Article14. Lally RT, Ederer MN, Woolfrey BF. Evaluation of mannitol salt agar with oxacillin as a screening medium for methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 1985. 22:501–504.
Article15. Clinical and Laboratory Standards Institute. CLSI document M100-S19. Performance standards for antimicrobial susceptibility testing nineteenth informational supplement. 2009. Wayne PA: Clinical and Laboratory Standards Institute.16. Schwartz DC, Cantor CR. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984. 37:67–75.
Article17. Murchan S, Kaufmann ME, Deplano A, deRyck R, Struelens M, Zinn CE, Fussing V, Salmenlinna S, Vuopio-Varkila J, El Solh N, Cuny C, Witte W, Tassios PT, Legakis N, van Leeuwen W, van Belkum A, Vindel A, Laconcha I, Garaizar J, Haeggman S, Olsson-Liljequist B, Ransjo U, Coombes G, Cookson B. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J Clin Microbiol. 2003. 41:1574–1585.
Article18. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995. 33:2233–2239.
Article19. Cha HY, Moon DC, Choi CH, Oh JY, Jeong YS, Lee YC, Seol SY, Cho DT, Chang HH, Kim SW, Lee JC. Prevalence of the ST239 clone of methicillin-resistant Staphylococcus aureus and differences in antimicrobial susceptibilities of ST239 and ST5 clones identified in a Korean hospital. J Clin Microbiol. 2005. 43:3610–3614.
Article20. Handley SA, Regnery RL. Differentiation of pathogenic Bartonella species by infrequent.restriction-site PCR. J Clin Microbiol. 2000. 38:3010–3015.
Article21. Dyer DW, Iandolo JJ. Rapid isolation of DNA from Staphylococcus aureus. Appl Environ Microbiol. 1983. 46:283–295.22. Riffard S, Lo Presti F, Vandenesch F, Forey F, Reyrolle M, Etienne J. Comparative analysis of infrequent-restriction-site PCR and pulsed-field gel electrophoresis for epidemiological typing of Legionella pneumophila serogroup 1 strains. J Clin Microbiol. 1998. 36:161–167.
Article23. Kim SI, Yoo JH, Cho YK, Lee DG, Wie SH, Choi JH, Kim YR, Shin WS, Kang MW. Comparison of Pulsed-field Gel Electrophoresis, Amplified Fragment Length Polymorphism, and Infrequent Restriction Site-Polymerase Chain Reaction for Molecular Typing of Escherichia coli and Staphylococcus aureus Strains. Korean J Infect Dis. 1999. 31:474–480.24. Yoo JH, Choi JH, Shin WS, Huh DH, Cho YK, Kim KM, Kim MY, Kang MW. Application of infrequent-restriction-site PCR to clinical isolates of Acinetobacter baumannii and Serratia marcescens. J Clin Microbiol. 1999. 37:3108–3112.
Article25. Peacock SJ, de Silva I, Lowy FD. What determines nasal carriage of Staphylococcus aureus? Trends Microbiol. 2001. 9:605–610.26. Fridkin SK, Hageman JC, Morrison M, Sanza LT, Como-Sabetti K, Jernigan JA, Harriman K, Harrison LH, Lynfield R, Farley MM. Active Bacterial Core Surveillance Program of the Emerging Infections Program Network. Methicillin-resistant Staphylococcus aureus disease in three communities. N Engl J Med. 2005. 352:1436–1444.
Article27. Hisata K, Kuwahara-Arai K, Yamanoto M, Ito T, Nakatomi Y, Cui L, Baba T, Terasawa M, Sotozono C, Kinoshita S, Yamashiro Y, Hiramatsu K. Dissemination of methicillin-resistant staphylococci among healthy Japanese children. J Clin Microbiol. 2005. 43:3364–3372.
Article28. Garner JS, Jarvis WR, Emori TG, Horan TC, Hughes JM. CDC definitions for nosocomial infections, 1988. Am J Infect Control. 1988. 16:128–140.
Article29. Lee WG. Resistance mechanism and epidermiology of vancomycin resistant enterococci. Korean J Clin Microbiol. 2008. 11:71–77.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Molecular Epidemiologic Methods Used in the Analysis of Methicillin-Resistant staphylococcus aureus
- Comparison of Pulsed-field Gel Electrophoresis, Amplified Fragment Length Polymorphism, and Infrequent Restriction Site-Polymerase Chain Reaction for Molecular Typing of Escherichia coli and Staphylococcus aureus Strains
- Comparison of infrequent restriction site-polymerase chain reaction and pulsed-field gel electrophoresis for molecular typing of Staphylococcus aureus and Escherichia coli
- Molecular Epidemiology of Methicillin-resistant Staphylococcus aureus Outbreak by Plasmid Restriction Analysis
- Molecular Epidemiologic Analysis of Community-Onset Extended Spectrum Beta-Lactamase (ESBL) Producing Escherichia coli Using Infrequent-Restriction-Site Polymerase Chain Reaction (IRS-PCR) with Comparison by Pulsed-Field Gel Electrophoresis (PFGE)