J Gynecol Oncol.
2012 Jul;23(3):182-189. 10.3802/jgo.2012.23.3.182.
The relationship between cisplatin resistance and histone deacetylase isoform overexpression in epithelial ovarian cancer cell lines
- Affiliations
-
- 1Department of Obstetrics and Gynecology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea. hyeokkim@amc.seoul.kr
- 2Asan Institute for Life Sciences, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea.
- KMID: 2245177
- DOI: http://doi.org/10.3802/jgo.2012.23.3.182
Abstract
OBJECTIVE
To investigate the relationship between cisplatin resistance and histone deacetylase (HDAC) isoform overexpression in ovarian cancer cell lines.
METHODS
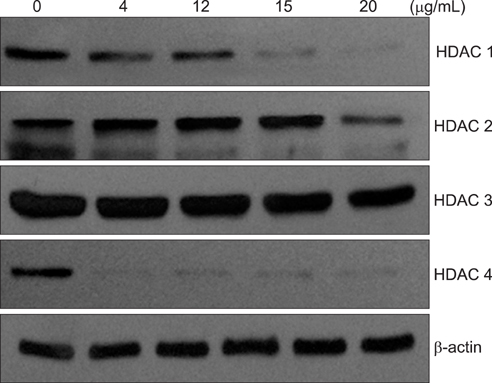
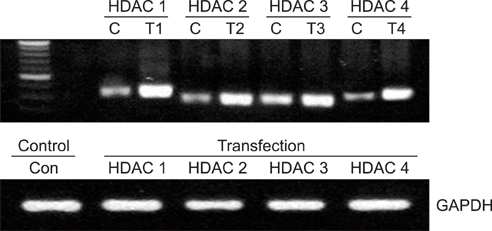
Expression of four HDAC isoforms (HDAC 1, 2, 3, and 4) in two ovarian cancer cell lines, SKOV3 and OVCAR3, exposed to various concentrations of cisplatin was examined by western blot analyses. Cells were transfected with plasmid DNA of each HDAC. The overexpression of protein and mRNA of each HDAC was confirmed by western blot and reverse transcriptase-polymerase chain reaction analyses, respectively. The cell viability of the SKOV3 and OVCAR3 cells transfected with HDAC plasmid DNA was measured using the cell counting kit-8 assay after treatment with cisplatin.
RESULTS
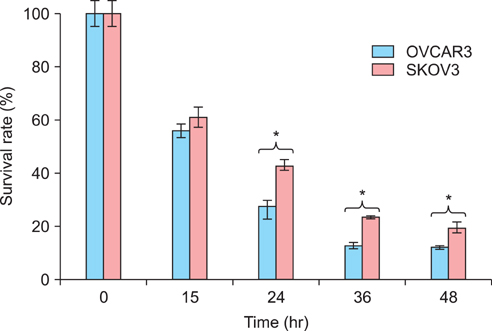
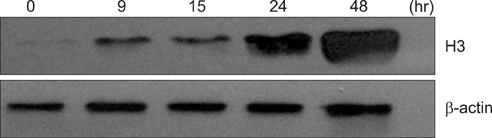
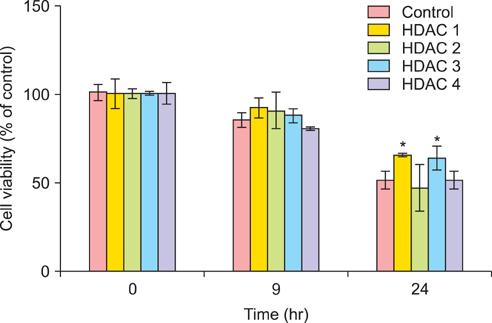
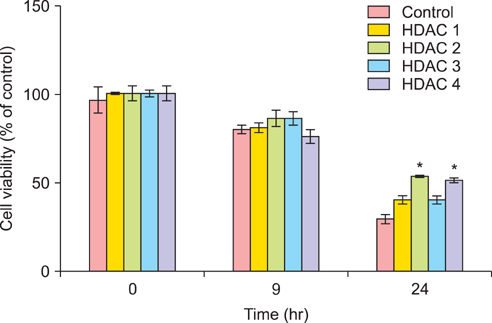
The 50% inhibitory concentration of the SKOV3 and OVCAR3 cells can be determined 15-24 hours after treatment with 15 microg/mL cisplatin. The expression level of acetylated histone 3 protein in SKOV3 cells increased after exposure to cisplatin. Compared with control cells at 24 hours after cisplatin exposure, the viability of SKOV3 cells overexpressing HDAC 1 and 3 increased by 15% and 13% (p<0.05), respectively. On the other hand, OVCAR3 cells that overexpressed HDAC 2 and 4 exhibited increased cell viability by 23% and 20% (p<0.05), respectively, compared with control cells 24 hours after exposure to cisplatin.
CONCLUSION
In SKOV3 and OVCAR3 epithelial ovarian cancer cell lines, the correlation between HDAC overexpression and cisplatin resistance was confirmed. However, the specific HDAC isoform associated with resistance to cisplatin varied depending on the ovarian cancer cell line. These results may suggest that each HDAC isoform conveys cisplatin resistance via different mechanisms.
MeSH Terms
-
Blotting, Western
Cell Count
Cell Line
Cell Survival
Cisplatin
DNA
Hand
Histone Deacetylases
Histones
Neoplasms, Glandular and Epithelial
Ovarian Neoplasms
Plasmids
Protein Isoforms
RNA, Messenger
Cisplatin
DNA
Histone Deacetylases
Histones
Neoplasms, Glandular and Epithelial
Ovarian Neoplasms
Protein Isoforms
RNA, Messenger
Figure
Reference
-
1. Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int J Cancer. 2001. 94:153–156.2. Ozols RF. Treatment goals in ovarian cancer. Int J Gynecol Cancer. 2005. 15:Suppl 1. 3–11.3. Chen Z, Clark S, Birkeland M, Sung CM, Lago A, Liu R, et al. Induction and superinduction of growth arrest and DNA damage gene 45 (GADD45) alpha and beta messenger RNAs by histone deacetylase inhibitors trichostatin A (TSA) and butyrate in SW620 human colon carcinoma cells. Cancer Lett. 2002. 188:127–140.4. Sato N, Ohta T, Kitagawa H, Kayahara M, Ninomiya I, Fushida S, et al. FR901228, a novel histone deacetylase inhibitor, induces cell cycle arrest and subsequent apoptosis in refractory human pancreatic cancer cells. Int J Oncol. 2004. 24:679–685.5. Yamashita Y, Shimada M, Harimoto N, Rikimaru T, Shirabe K, Tanaka S, et al. Histone deacetylase inhibitor trichostatin A induces cell-cycle arrest/apoptosis and hepatocyte differentiation in human hepatoma cells. Int J Cancer. 2003. 103:572–576.6. Kelly WK, Richon VM, O'Connor O, Curley T, MacGregor-Curtelli B, Tong W, et al. Phase I clinical trial of histone deacetylase inhibitor: suberoylanilide hydroxamic acid administered intravenously. Clin Cancer Res. 2003. 9:3578–3588.7. Edwards A, Li J, Atadja P, Bhalla K, Haura EB. Effect of the histone deacetylase inhibitor LBH589 against epidermal growth factor receptor-dependent human lung cancer cells. Mol Cancer Ther. 2007. 6:2515–2524.8. Fuino L, Bali P, Wittmann S, Donapaty S, Guo F, Yamaguchi H, et al. Histone deacetylase inhibitor LAQ824 down-regulates Her-2 and sensitizes human breast cancer cells to trastuzumab, taxotere, gemcitabine, and epothilone B. Mol Cancer Ther. 2003. 2:971–984.9. Kell J. Drug evaluation: MGCD-0103, a histone deacetylase inhibitor for the treatment of cancer. Curr Opin Investig Drugs. 2007. 8:485–492.10. Saito A, Yamashita T, Mariko Y, Nosaka Y, Tsuchiya K, Ando T, et al. A synthetic inhibitor of histone deacetylase, MS-27-275, with marked in vivo antitumor activity against human tumors. Proc Natl Acad Sci U S A. 1999. 96:4592–4597.11. Sandor V, Bakke S, Robey RW, Kang MH, Blagosklonny MV, Bender J, et al. Phase I trial of the histone deacetylase inhibitor, depsipeptide (FR901228, NSC 630176), in patients with refractory neoplasms. Clin Cancer Res. 2002. 8:718–728.12. Jin KL, Pak JH, Park JY, Choi WH, Lee JY, Kim JH, et al. Expression profile of histone deacetylases 1, 2 and 3 in ovarian cancer tissues. J Gynecol Oncol. 2008. 19:185–190.13. Lengauer C, Issa JP. The role of epigenetics in cancer. DNA Methylation, Imprinting and the Epigenetics of Cancer: An American Association for Cancer Research Special Conference. Las Croabas, Puerto Rico, 12-16 1997 December. Mol Med Today. 1998. 4:102–103.14. Muscolini M, Cianfrocca R, Sajeva A, Mozzetti S, Ferrandina G, Costanzo A, et al. Trichostatin A up-regulates p73 and induces Bax-dependent apoptosis in cisplatin-resistant ovarian cancer cells. Mol Cancer Ther. 2008. 7:1410–1419.15. Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002. 3:415–428.16. Takacs M, Salamon D, Myohanen S, Li H, Segesdi J, Ujvari D, et al. Epigenetics of latent Epstein-Barr virus genomes: high resolution methylation analysis of the bidirectional promoter region of latent membrane protein 1 and 2B genes. Biol Chem. 2001. 382:699–705.17. Zuccotti M, Garagna S, Redi CA. Nuclear transfer, genome reprogramming and novel opportunities in cell therapy. J Endocrinol Invest. 2000. 23:623–629.18. Howell CY, Bestor TH, Ding F, Latham KE, Mertineit C, Trasler JM, et al. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell. 2001. 104:829–838.19. Davie JR. Covalent modifications of histones: expression from chromatin templates. Curr Opin Genet Dev. 1998. 8:173–178.20. de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003. 370:737–749.21. Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004. 338:17–31.22. Yang QC, Zeng BF, Shi ZM, Dong Y, Jiang ZM, Huang J, et al. Inhibition of hypoxia-induced angiogenesis by trichostatin A via suppression of HIF-1a activity in human osteosarcoma. J Exp Clin Cancer Res. 2006. 25:593–599.23. Chen B, Cepko CL. HDAC4 regulates neuronal survival in normal and diseased retinas. Science. 2009. 323:256–259.24. Thiagalingam S, Cheng KH, Lee HJ, Mineva N, Thiagalingam A, Ponte JF. Histone deacetylases: unique players in shaping the epigenetic histone code. Ann N Y Acad Sci. 2003. 983:84–100.25. Bargonetti J, Manfredi JJ. Multiple roles of the tumor suppressor p53. Curr Opin Oncol. 2002. 14:86–91.26. Bandyopadhyay D, Mishra A, Medrano EE. Overexpression of histone deacetylase 1 confers resistance to sodium butyrate-mediated apoptosis in melanoma cells through a p53-mediated pathway. Cancer Res. 2004. 64:7706–7710.27. Basile V, Mantovani R, Imbriano C. DNA damage promotes histone deacetylase 4 nuclear localization and repression of G2/M promoters, via p53 C-terminal lysines. J Biol Chem. 2006. 281:2347–2357.28. Zeng L, Xiao Q, Margariti A, Zhang Z, Zampetaki A, Patel S, et al. HDAC3 is crucial in shear-and VEGF-induced stem cell differentiation toward endothelial cells. J Cell Biol. 2006. 174:1059–1069.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Relationship between Expression of 1-Cys Peroxiredoxin and Resistance to Cisplatin in Epithelial Ovarian Cancer Cell Lines
- Expression of p53, p16, PTEN, and c-myc Gene with Cisplatin Treatment in Cisplatin Resistant Ovarian Cancer Cell Line
- Epigenetic modification of α-N-acetylgalactosaminidase enhances cisplatin resistance in ovarian cancer
- Apoptosis-related mRNA expression profiles of ovarian cancer cell lines following cisplatin treatment
- SP1-induced lncRNA MCF2L-AS1 promotes cisplatin resistance in ovarian cancer by regulating IGF2BP1/IGF2/MEK/ERK axis