Korean J Lab Med.
2006 Aug;26(4):299-306. 10.3343/kjlm.2006.26.4.299.
Development of a Web-based Program to Calculate Sample Size for Evaluating the Performance of In Vitro Diagnostic Kits
- Affiliations
-
- 1Department of Laboratory Medicine, Pusan National University Hospital, Pusan National University School of Medicine, Busan, Korea.
- 2Department of Laboratory Medicine, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea. hboh@amc.seoul.kr
- 3Department of Computer Science Education, Korea University, Seoul, Korea.
- 4Department of Statistics, Dongkuk University, Seoul, Korea.
- KMID: 2238839
- DOI: http://doi.org/10.3343/kjlm.2006.26.4.299
Abstract
-
BACKGROUND: Many studies evaluating the performance of in vitro diagnostic kits have been criticized for the lack of reliability. To attain reliability those evaluation studies should be preceded by sample size calculation ensuring statistical power. This study was intended to develop a web-based system to estimate the sample size, which was often neglected because it would require expert knowledge in statistics.
METHODS
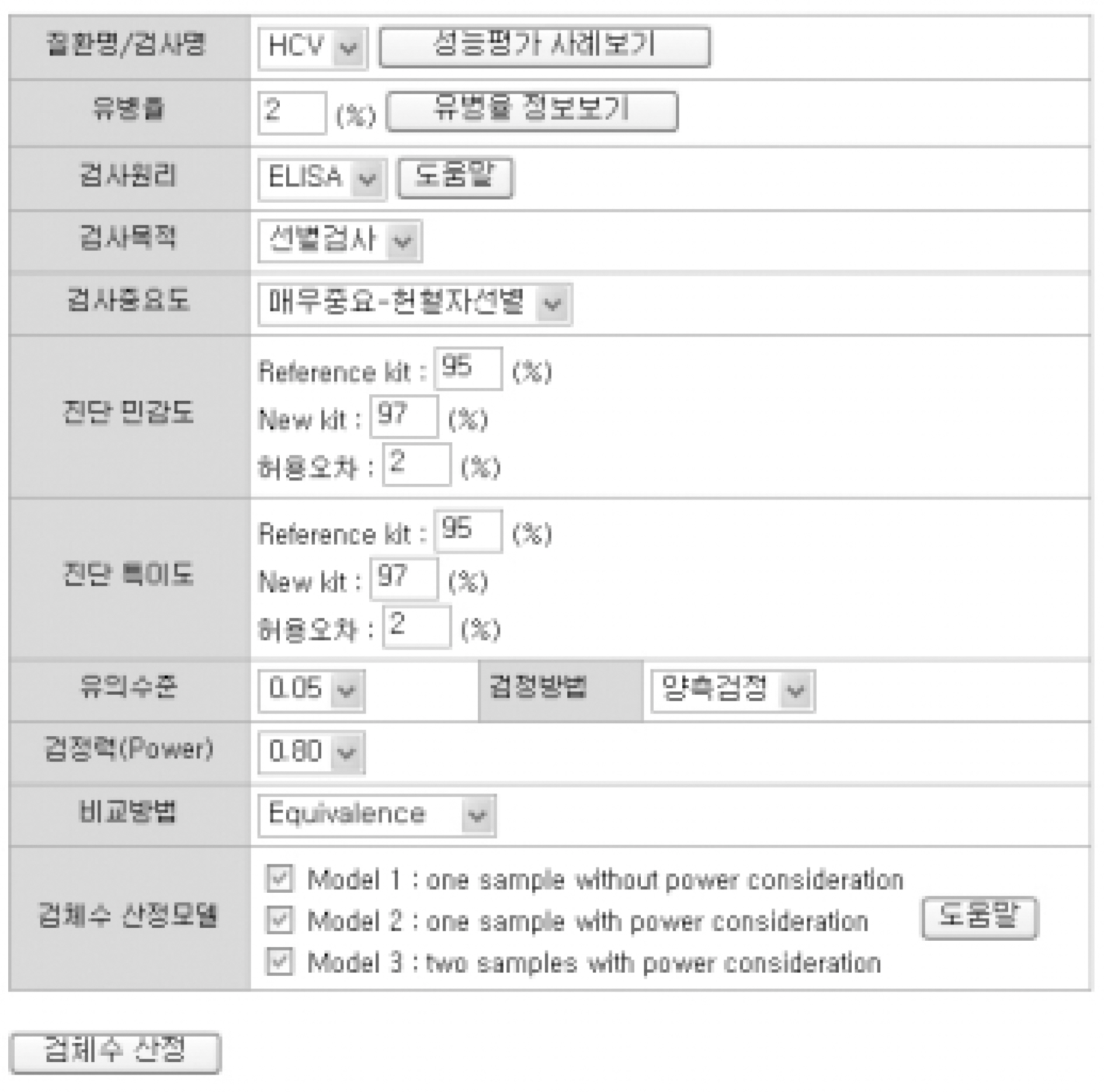
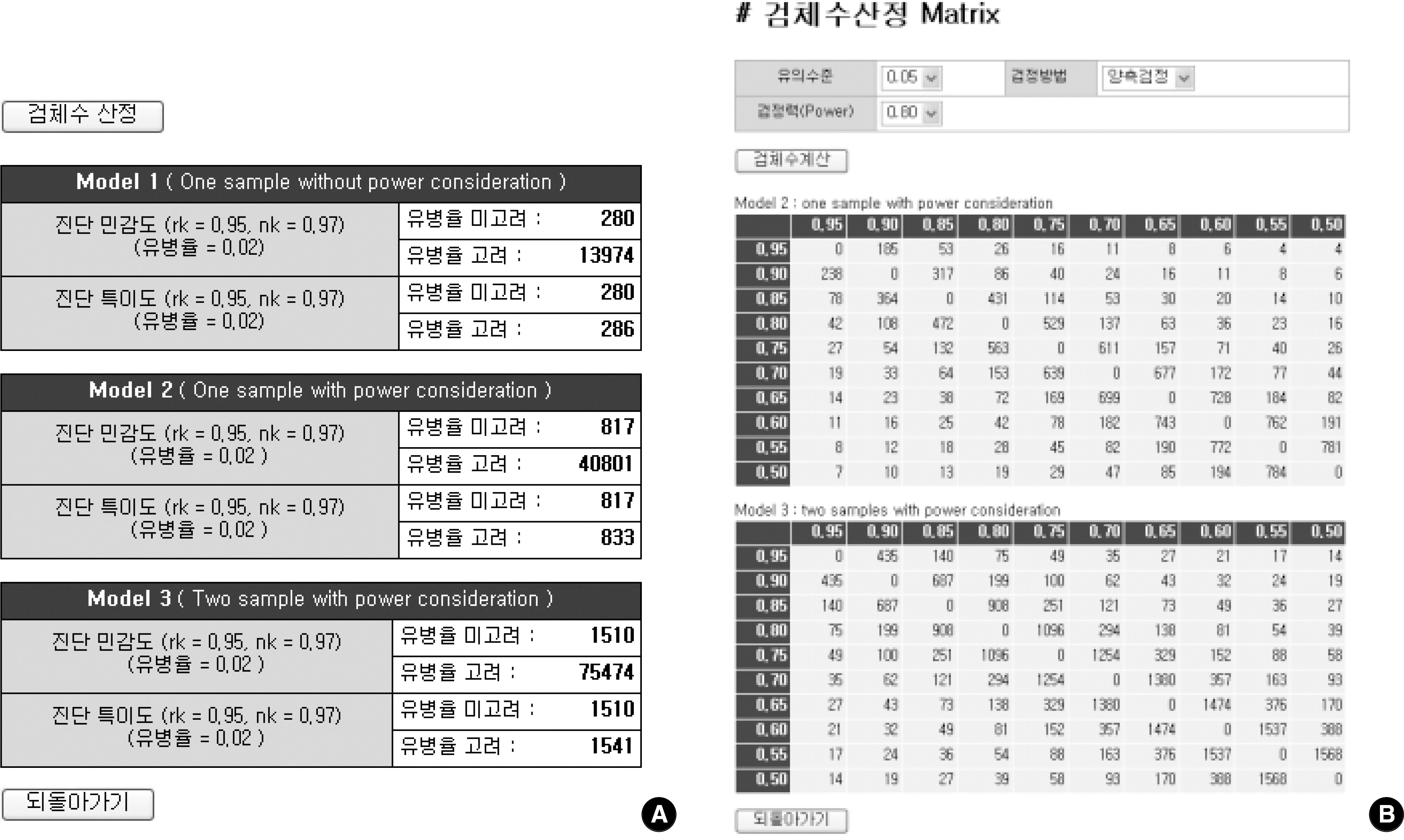
For sample size calculation, we extracted essential parameters from the performance studies on the 3rd generation anti-hepatitis C virus (HCV) kits reported in the literature. We developed a system with PHP web-script language and MySQL. The statistical models used in this system were as follows; one sample without power consideration (model 1), one sample with power consideration (model 2), and two samples with power consideration (model 3).
RESULTS
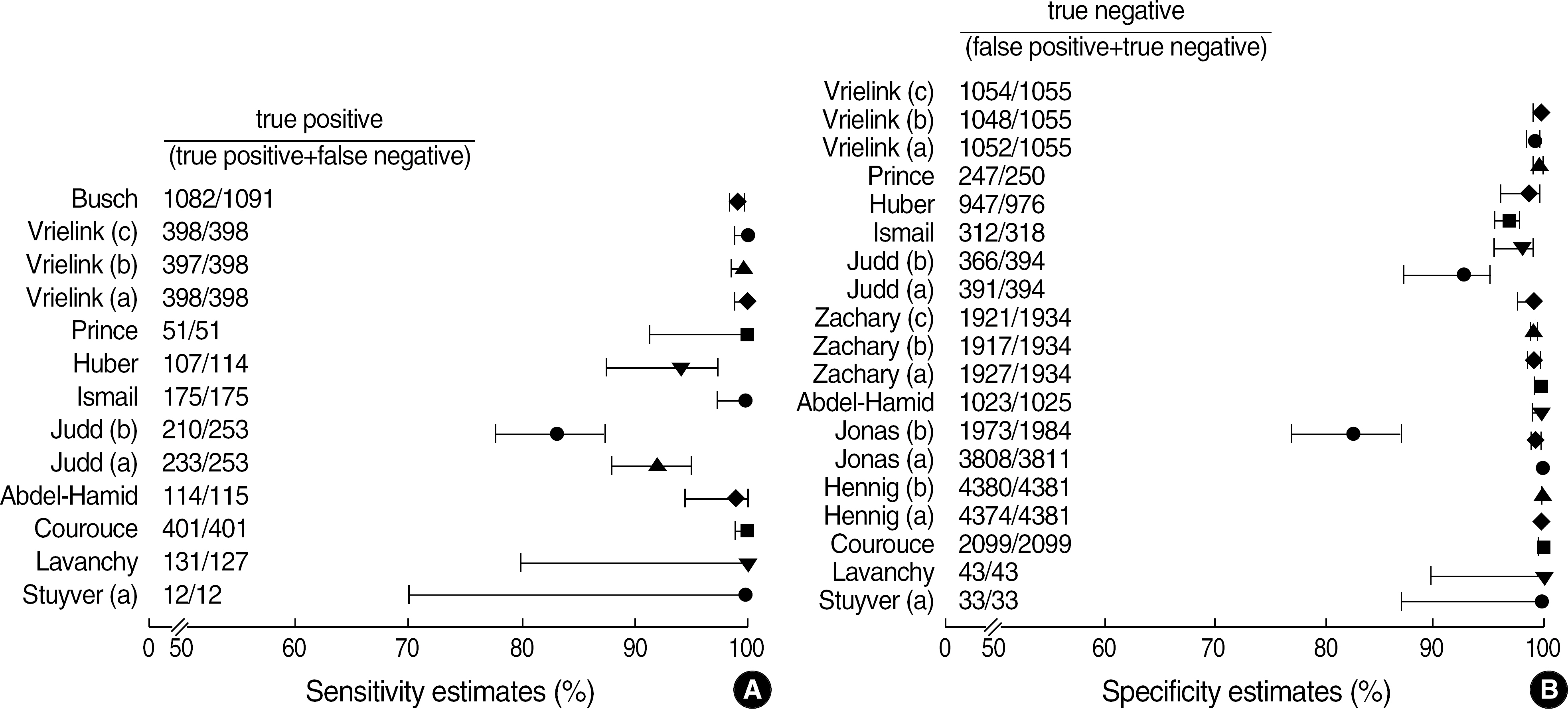
Among the articles published between 1989 and 2005, 13 articles that evaluated the performance of anti-HCV kits were identified by searching with Medical Subject Headings (MeSH). The diagnostic sensitivity was 83-100% with a median of 145 samples (range; 12-1,091) and the specificity was 97-100% with a median of 1,025 samples (range; 33-4,381). The estimated sample size would be 280 in the model 1, 817 in the model 2, and 1,510 in the model 3, when we set 2% prevalence of HCV infection, 95% sensitivity of a conventional kit, 97% sensitivity of a new kit , 95% significance level (two-sided test), 2% allowable error, and 80% power.
CONCLUSIONS
Our study indicates that an insufficient sample size is still a problem in performance evaluation. Our system should be helpful in increasing the reliability of performance evaluation by providing an appropriate sample size.
Keyword
MeSH Terms
Figure
Reference
-
References
1. How to read clinical journals: II. To learn about a diagnostic test. Can Med Assoc J. 1981; 124:703–10.2. Deeks JJ, Altman DG. Diagnostic tests 4: likelihood ratios. BMJ. 2004; 329:168–9.
Article3. Sim J, Wright CC. The kappa statistic in reliability studies: use, interpretation, and sample size requirements. Phys Ther. 2005; 85:257–68.
Article4. Deeks JJ. Systematic reviews in health care: Systematic reviews of evaluations of diagnostic and screening tests. BMJ. 2001; 323:157–62.
Article5. Reid MC, Lachs MS, Feinstein AR. Use of methodological standards in diagnostic test research. Getting better but still not good. JAMA. 1995; 274:645–51.
Article6. Lumbreras-Lacarra B, Ramos-Rincon JM, Hernandez-Aguado I. Methodology in diagnostic laboratory test research in clinical chemistry and clinical chemistry and laboratory medicine. Clin Chem. 2004; 50:530–6.
Article7. Freiman JA, Chalmers TC, Smith H Jr, Kuebler RR. The importance of beta, the type II error and sample size in the design and interpretation of the randomized control trial. Survey of 71 “negative” trials. N Engl J Med. 1978; 299:690–4.8. Moore AD, Joseph L. Sample size considerations for superiority trials in systemic lupus erythematosus (SLE). Lupus. 1999; 8:612–9.9. Newcombe RG. Two-sided confidence intervals for the single proportion: comparison of seven methods. Stat Med. 1998; 17:857–72.
Article10. Arkin CF, Wachtel MS. How many patients are necessary to assess test performance? JAMA. 1990; 263:275–8.
Article11. Wang JT, Wang TH, Sheu JC, Tsai SJ, Hsieh YS, Lin DT, et al. Hepatitis C virus infection in volunteer blood donors in Taiwan. Evaluation by hepatitis C antibody assays and the polymerase chain reaction. Arch Pathol Lab Med. 1993; 117:152–6.12. Colin C, Lanoir D, Touzet S, Meyaud-Kraemer L, Bailly F, Trepo C. HEPATITIS Group. Sensitivity and specificity of third-generation hepatitis C virus antibody detection assays: an analysis of the literature. J Viral Hepat. 2001; 8:87–95.
Article13. Dati F, Denoyel G, van Helden J. European performance evaluations of the ADVIA Centaur infectious disease assays: requirements for performance evaluation according to the European directive on in vitro diagnostics. J Clin Virol. 2004; 30:S6–10.
Article14. Rutjes AW, Reitsma JB, Vandenbroucke JP, Glas AS, Bossuyt PM. Case-control and two-gate designs in diagnostic accuracy studies. Clin Chem. 2005; 51:1335–41.
Article15. Sackett DL, Haynes RB. The architecture of diagnostic research. BMJ. 2002; 324:539–41.16. Gluud C, Gluud LL. Evidence based diagnostics. BMJ. 2005; 330:724–6.
Article17. Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig LM, et al. Towards complete and accurate reporting of studies of diagnostic accuracy: The STARD Initiative. Ann Intern Med. 2003; 138:40–4.
Article18. Smith BJ, Darzins PJ, Quinn M, Heller RF. Modern methods of searching the medical literature. Med J Aust. 1992; 157:603–11.19. Deville WL, Bezemer PD, Bouter LM. Publications on diagnostic test evaluation in family medicine journals: an optimal search strategy. J Clin Epidemiol. 2000; 53:65–9.20. Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig LM, et al. The STARD statement for reporting studies of diagnostic accuracy: explanation and elaboration. Ann Intern Med. 2003; 138:1–12.
Article21. Stuyver L, Claeys H, Wyseur A, Van Arnhem W, De Beenhouwer H, Uytendaele S, et al. Hepatitis C virus in a hemodialysis unit: molecular evidence for nosocomial transmission. Kidney Int. 1996; 49:889–95.
Article22. Lavanchy D, Steinmann J, Moritz A, Frei PC. Evaluation of a new automated third-generation anti-HCV enzyme immunoassay. J Clin Lab Anal. 1996; 10:269–76.
Article23. Courouce AM, Bouchardeau F, Girault A, Le Marrec N. Significance of NS3 and NS5 antigens in screening for HCV antibody. Lancet. 1994; 343:853–4.24. Hennig H, Schlenke P, Kirchner H, Bauer I, Schulte-Kellinghaus B, Bludau H. Evaluation of newly developed microparticle enzyme immunoassays for the detection of HCV antibodies. J Virol Methods. 2000; 84:181–90.
Article25. Jonas G, Pelzer C, Beckert C, Hausmann M, Kapprell HP. Performance characteristics of the ARCHITECT anti-HCV assay. J Clin Virol. 2005; 34:97–103.
Article26. Abdel-Hamid M, El-Daly M, El-Kafrawy S, Mikhail N, Strickland GT, Fix AD. Comparison of second- and third-generation enzyme immunoassays for detecting antibodies to hepatitis C virus. J Clin Microbiol. 2002; 40:1656–9.
Article27. Zachary P, Ullmann M, Djeddi S, Wendling MJ, Schvoerer E, Stoll-Keller F, et al. Evaluation of two commercial enzyme immunoassays for diagnosis of hepatitis C in the conditions of a virology laboratory. Pathol Biol. 2004; 52:511–6.28. Judd A, Parry J, Hickman M, McDonald T, Jordan L, Lewis K, et al. Evaluation of a modified commercial assay in detecting antibody to hepatitis C virus in oral fluids and dried blood spots. J Med Virol. 2003; 71:49–55.
Article29. Ismail N, Fish GE, Smith MB. Laboratory evaluation of a fully automated chemiluminescence immunoassay for rapid detection of HBsAg, antibodies to HBsAg, and antibodies to hepatitis C virus. J Clin Microbiol. 2004; 42:610–7.
Article30. Huber KR, Sebesta C, Bauer K. Detection of common hepatitis C virus subtypes with a third-generation enzyme immunoassay. Hepatology. 1996; 24:471–3.31. Prince AM, Scheffel JW, Moore B. A search for hepatitis C virus polymerase chain reaction-positive but seronegative subjects among blood donors with elevated alanine aminotransferase. Transfusion. 1997; 37:211–4.
Article32. Vrielink H, Zaaijer HL, Reesink HW, van der Poel CL, Cuypers HT, Lelie PN. Sensitivity and specificity of three third-generation anti-hepatitis C virus ELISAs. Vox Sang. 1995; 69:14–7.
Article33. Busch MP, Watanabe KK, Smith JW, Hermansen SW, Thomson RA. False-negative testing errors in routine viral marker screening of blood donors. For the Retrovirus Epidemiology Donor Study. Transfusion. 2000; 40:585–9.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Development and Evaluation of a Web-based Education Program for Nursing Students on Control of Vancomycin-resistant Enterococcus Infection
- Considerations when calculating the sample size for an inequality test
- Development and Application of a Web-Based Tailored Nutrition Management Program and Change in Knowledge of Nutrition and Eating Habits in Elementary Students
- Development and Evaluation of a Web-based Support Program for the Maternal Role of Primiparas
- How to Calculate Sample Size and Why