J Bacteriol Virol.
2016 Mar;46(1):22-26. 10.4167/jbv.2016.46.1.22.
Application of Diagnostic Microarray Technique in Subtyping and Pathotyping of Avian Influenza Viruses Isolated in Mongolia
- Affiliations
-
- 1Avian Disease Laboratory, College of Veterinary Medicine, Konkuk University, Seoul, Korea. songcs@konkuk.ac.kr
- 2Bio-Core Co. Ltd., Seoul, Korea.
- 3Department of Molecular and Life Science, Hanyang University, Ansan, Gyeonggi-do, Korea.
- 4Department of Bio-Nanotechnology, Hanyang University, Gyeonggi-do, Korea.
- KMID: 2168739
- DOI: http://doi.org/10.4167/jbv.2016.46.1.22
Abstract
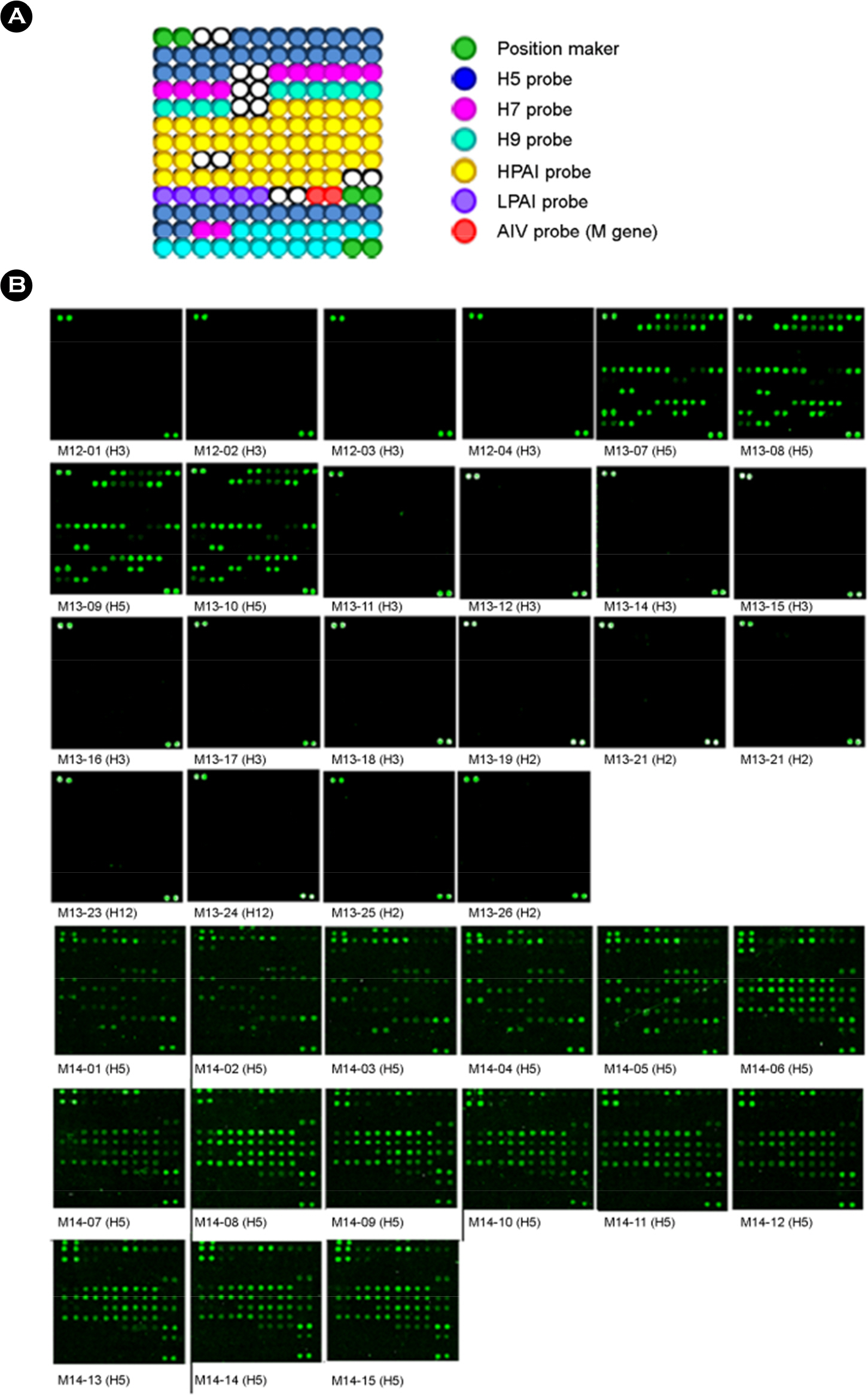
- Asian-lineage H5 highly pathogenic avian influenza (HPAI) viruses have caused continuous outbreaks in poultry and wild birds. Development of rapid and accurate diagnostic methods is needed for preventing further spread of the virus and reducing the time required for eradication of the virus. We developed a low-density microarray for the rapid detection and identification of avian influenza virus subtypes H5, H7, and H9 and their pathotypes in a previous study. In the present study, we evaluated previously developed diagnostic microarray using avian influenza viruses isolated in Mongolia, including H5 HPAI viruses. All H5 HPAI viruses isolated in Mongolia were shown as H5-specific and highly pathogenic pattern in the microarray. H2, H3 and H12 viruses isolated in Mongolia used in this study did not show any H5, H7 and H9 patterns. These results indicated that this diagnostic microarray has enormous potential for the rapid subtyping and pathotyping of influenza viruses, including viruses isolated in Mongolia.
Keyword
Figure
Reference
-
1). Sonnberg S, Webby RJ, Webster RG. Natural history of highly pathogenic avian influenza H5N1. Virus Res. 2013; 178:63–77.
Article2). Swayne DE, Pavade G, Hamilton K, Vallat B, Miyagishima K. Assessment of national strategies for control of high-pathogenicity avian influenza and low-pathogenicity notifiable avian influenza in poultry, with emphasis on vaccines and vaccination. Rev Sci Tech. 2011; 30:839–70.
Article3). Spackman E, Senne DA, Bulaga LL, Myers TJ, Perdue ML, Garber LP, et al. Development of real-time RT-PCR for the detection of avian influenza virus. Avian Dis. 2003; 47:1079–82.
Article4). Yoo SM, Choi JH, Lee SY, Yoo NC. Applications of DNA microarray in disease diagnostics. J Microbiol Biotechnol. 2009; 19:635–46.5). Lee DH, Chung IH, Roh MR, Lee HJ, Lee YJ, Jung JW, et al. A Diagnostic Microarray for Subtyping and Pathotyping Avian Influenza Virus. Biochip J. 2009; 3:57–64.6). Lee DH, Kim JH, Lee YN, Park JK, Yuk SS, Jung JW, et al. Simultaneous subtyping and pathotyping of the 2010–2011 South Korean HPAI outbreak strain by using a diagnostic microarray. Biochip J. 2011; 5:369–74.
Article7). Lee DH, Kim JH, Yuk SS, Kwon JH, Cho H, Hwang SY, et al. Rapid hemagglutinin subtyping of novel avian-origin influenza A (H7N9) virus using a diagnostic microarray. Biochip J. 2014; 8:55–9.8). Lee YJ, Kang HM, Lee EK, Song BM, Jeong J, Kwon YK, et al. Novel reassortant influenza A(H5N8) viruses, South Korea, 2014. Emerg Infect Dis. 2014; 20:1087–9.
Article9). Zhao G, Gu X, Lu X, Pan J, Duan Z, Zhao K, et al. Novel reassortant highly pathogenic H5N2 avian influenza viruses in poultry in China. PLoS One. 2012; 7:e46183.
Article10). Zhao K, Gu M, Zhong L, Duan Z, Zhang Y, Zhu Y, et al. Characterization of three H5N5 and one H5N8 highly pathogenic avian influenza viruses in China. Vet Microbiol. 2013; 163:351–7.
Article11). Sakoda Y, Sugar S, Batchluun D, Erdene-Ochir TO, Okamatsu M, Isoda N, et al. Characterization of H5N1 highly pathogenic avian influenza virus strains isolated from migratory waterfowl in Mongolia on the way back from the southern Asia to their northern territory. Virology. 2010; 406:88–94.
Article12). Kang HM, Batchuluun D, Kim MC, Choi JG, Erdene-Ochir TO, Paek MR, et al. Genetic analyses of H5N1 avian influenza virus in Mongolia, 2009 and its relationship with those of eastern Asia. Vet Microbiol. 2011; 147:170–5.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- H5 and H9 subtypes of Avian Influenza Viruses are Real Threat To Humans
- The significance of avian influenza virus mouse-adaptation and its application in characterizing the efficacy of new vaccines and therapeutic agents
- Surveillance of wild birds for avian influenza virus in Korea
- Pathogenicity of H5N8 virus in chickens from Korea in 2014
- Novel reassortants of clade 2.3.4.4 H5N6 highly pathogenic avian influenza viruses possessing genetic heterogeneity in South Korea in late 2017