J Bacteriol Virol.
2014 Jun;44(2):188-196. 10.4167/jbv.2014.44.2.188.
Genome Size Constraint in Replication and Packaging of Turnip Yellow Mosaic Virus
- Affiliations
-
- 1Department of Biochemistry, Chungbuk National University, Cheongju, Korea. tjcho@chungbuk.ac.kr
- KMID: 2168694
- DOI: http://doi.org/10.4167/jbv.2014.44.2.188
Abstract
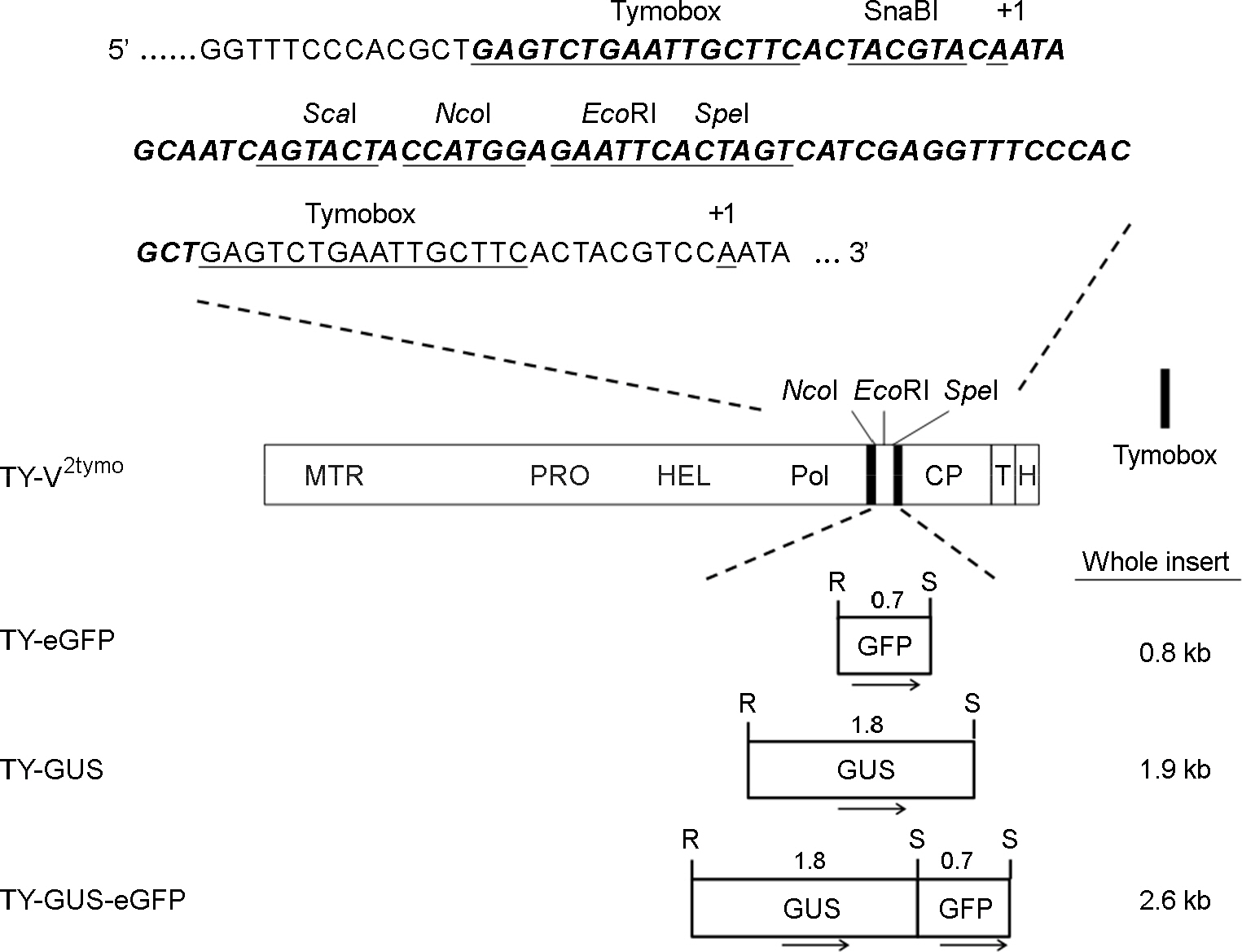
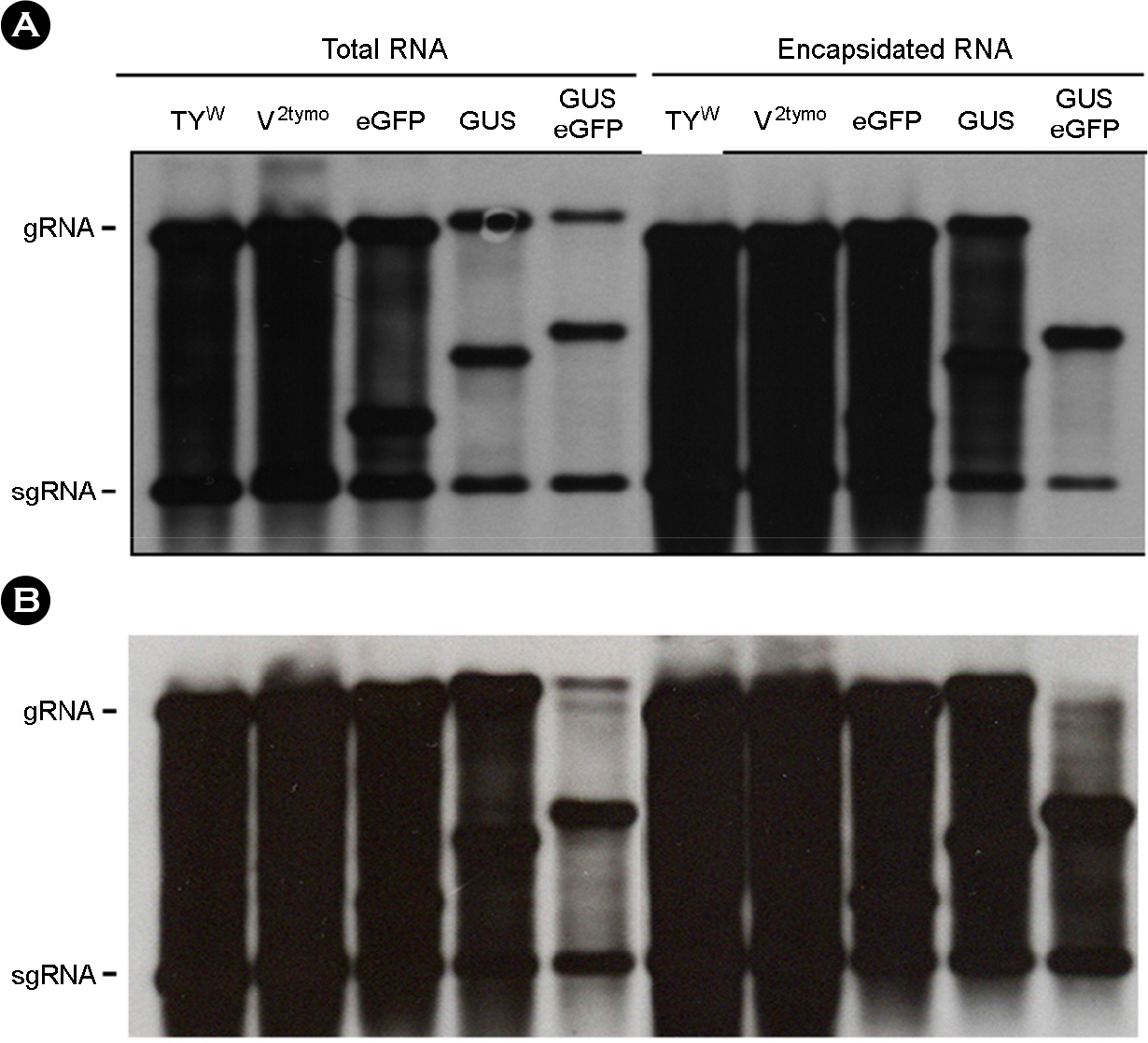
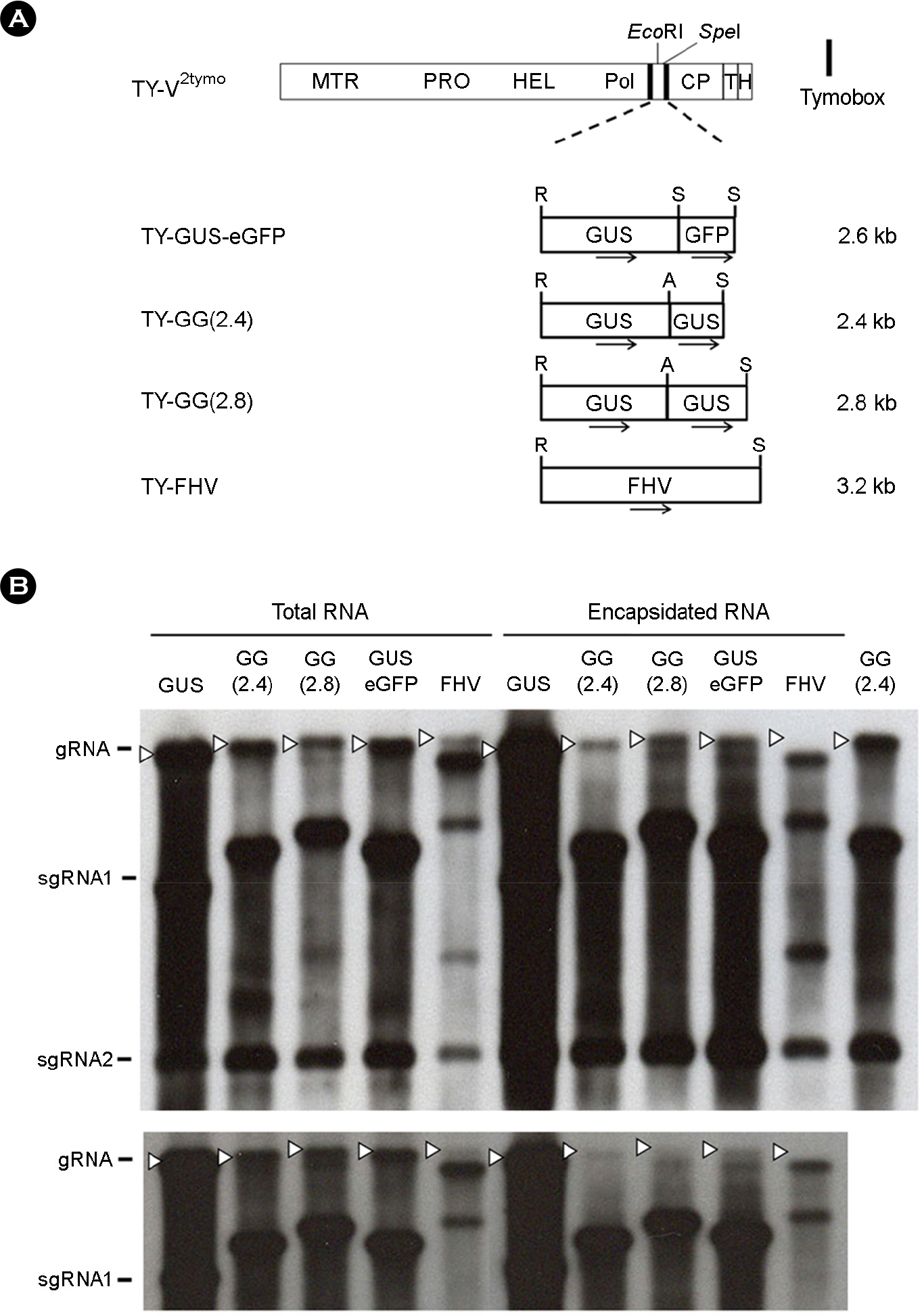
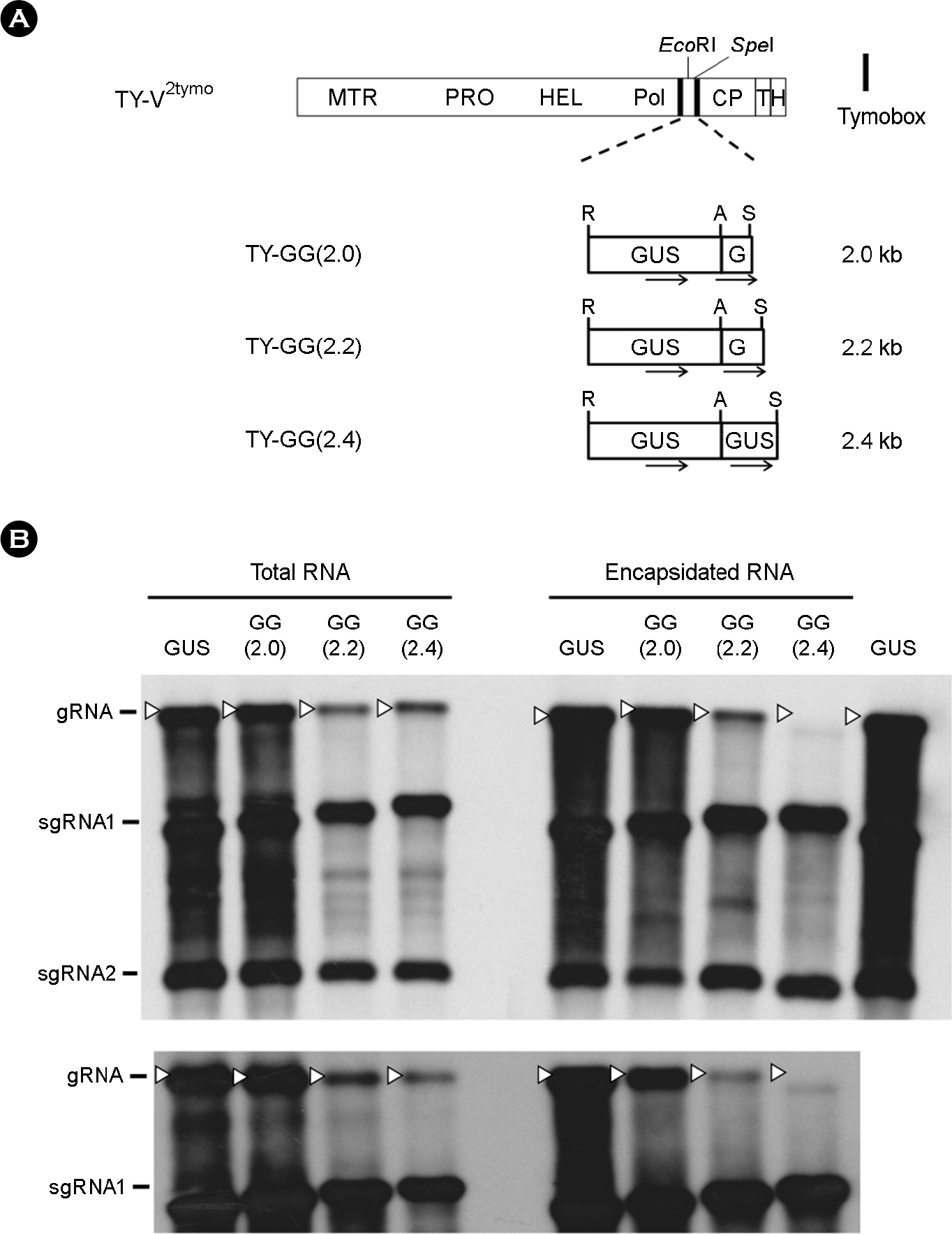
- Turnip yellow mosaic virus (TYMV) is a spherical plant virus that has a single 6.3 kb positive strand RNA as a genome. Previously, we have made the recombinant TYMV construct containing a 0.7 kb eGFP gene or a 1.8 kb GUS gene. The genomic RNAs from these constructs were efficiently encapsidated. To examine in more detail whether size constraint exists for replication and packaging of TYMV, we have inserted into the TY-GUS an extra sequence derived from either eGFP or GUS. We also made a recombinant containing RNA1 sequence of Flock house virus. These TYMV recombinants were introduced into Nicotiana benthamiana leaves by agroinfiltration. Northern blot analysis of the viral RNAs in the agroinfiltrated leaves showed that the genomic RNA band from the recombinant TYMV became weaker as longer sequence was inserted. The result also showed that the efficiency of genomic RNA encapsidation decreased sharply when an extra sequence of 2.2 kb or more was inserted. In contrast, the recombinant subgenomic RNA containing an extra sequence of up to 3.2 kb was efficiently encapsidated. Overall, these results show that size constraint exists for replication and encapsidation of TYMV RNA.
Keyword
MeSH Terms
Figure
Cited by 1 articles
-
N-terminal Extension of Coat Protein of Turnip Yellow Mosaic Virus has Variable Effects on Replication, RNA Packaging, and Virion Assembly Depending on the Inserted Sequence
Kwang-Hee Chae, Doyeong Kim, Tae-Ju Cho
J Bacteriol Virol. 2016;46(1):13-21. doi: 10.4167/jbv.2016.46.1.13.
Reference
-
1). Dreher TW. Turnip yellow mosaic virus: transfer RNA mimicry, chloroplasts and a C-rich genome. Mol Plant Pathol. 2004; 5:367–75.
Article2). Canady MA, Larson SB, Day J, McPherson A. Crystal structure of turnip yellow mosaic virus. Nat Struct Biol. 1996; 3:771–81.
Article3). Mellema JR, Benicourt C, Haenni AL, Noort A, Pleij CW, Bosch L. Translational studies with turnip yellow mosaic virus RNAs isolated from major and minor virus particles. Virology. 1979; 96:38–46.
Article4). Rao ALN. Genome packaging by spherical plant RNA viruses. Annu Rev Phytopathol. 2006; 44:61–87.
Article5). Qu F, Morris TJ. Encapsidation of turnip crinkle virus is defined by a specific packaging signal and RNA size. J Virol. 1997; 71:1428–35.
Article6). Shin HI, Kim IC, Cho TJ. Replication and encapsidation of recombinant Turnip yellow mosaic virus RNA. BMB Rep. 2008; 41:739–44.
Article7). Cho TJ, Dreher TW. Encapsidation of genomic but not subgenomic turnip yellow mosaic virus RNAs by coat protein provided in trans. Virology. 2006; 356:126–35.8). Voinnet O, Rivas S, Mestre P, Baulcombe D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 2003; 33:949–56.9). Albarino CG, Price BD, Eckerle LD, Ball LA. Characterization and template properties of RNA dimers generated during Flock house virus RNA replication. Virology. 2001; 289:269–82.
Article10). Matthews REF. Tymovirus group. Plant Virology. 3rd ed.Academic Press;1991. p. p231–39.11). Hellendoorn K, Michiels PJ, Buitenhuis R, Pleij CW. Protonatable hairpins are conserved in the 5′-untranslated region of tymovirus RNAs. Nucl Acids Res. 1996; 24:4910–7.
Article12). Hellendoorn K, Verlaan PWG, Pleij CWA. A functional role for the conserved protonatable hairpins in the 5′ untranslated region of turnip yellow mosaic virus RNA. J Virol. 1997; 71:8774–9.
Article13). Bink HHJ, Hellendoorn K, van der Meulen J, Pleij CWA. Protonation of non-Watson-Crick base pairs and encapsidation of turnip yellow mosaic virus RNA. Proc Natl Acad Sci. 2002; 99:13465–70.
Article14). Shin HI, Tzanetakis IE, Dreher TW, Cho TJ. The 5′-UTR of Turnip yellow mosaic virus does not include a critical encapsidation signal. Virology. 2009; 387:427–35.
Article15). Choi YG, Dreher TW, Rao ALN. tRNA elements mediate the assembly of an icosahedral RNA virus. Proc Natl Acad Sci. 2002; 99:655–60.
Article16). Shin HI, Kim HY, Cho TJ. The Pro/Hel region is indispensable for packaging non-replicating Turnip yellow mosaic virus RNA, but not replicating viral RNA. Mol. Cells. 2010; 29:463–9.17). Shin HI, Cho NJ, Cho TJ. Role of 5′-UTR hairpins of the Turnip yellow mosaic virus RNA in replication and systemic movement. BMB Rep. 2008; 41:778–83.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A Sequence in Coat Protein Open Reading Frame Is Required for Turnip Yellow Mosaic Virus Replication
- N-terminal Extension of Coat Protein of Turnip Yellow Mosaic Virus has Variable Effects on Replication, RNA Packaging, and Virion Assembly Depending on the Inserted Sequence
- Replication of Recombinant Flock House Virus RNA Encapsidated by Turnip Yellow Mosaic Virus Coat Proteins in Nicotiana benthamiana
- Characterization of a Replication Element in the Coat Protein ORF of Turnip Yellow Mosaic Virus
- Read-through Mutation in the Coat Protein ORF Suppresses Turnip Yellow Mosaic Virus Subgenomic RNA Accumulation