J Bacteriol Virol.
2012 Dec;42(4):363-367. 10.4167/jbv.2012.42.4.363.
An Universal Approach to Getting Ahead for Influenza B Vaccines
- Affiliations
-
- 1Department of Microbiology, College of Medicine, Hallym University, Chuncheon, Gangwon-do, Korea. manseong.park@gmail.com
- 2Center for Medical Science Research, College of Medicine, Hallym University, Chuncheon, Gangwon-do, Korea.
- KMID: 2168674
- DOI: http://doi.org/10.4167/jbv.2012.42.4.363
Abstract
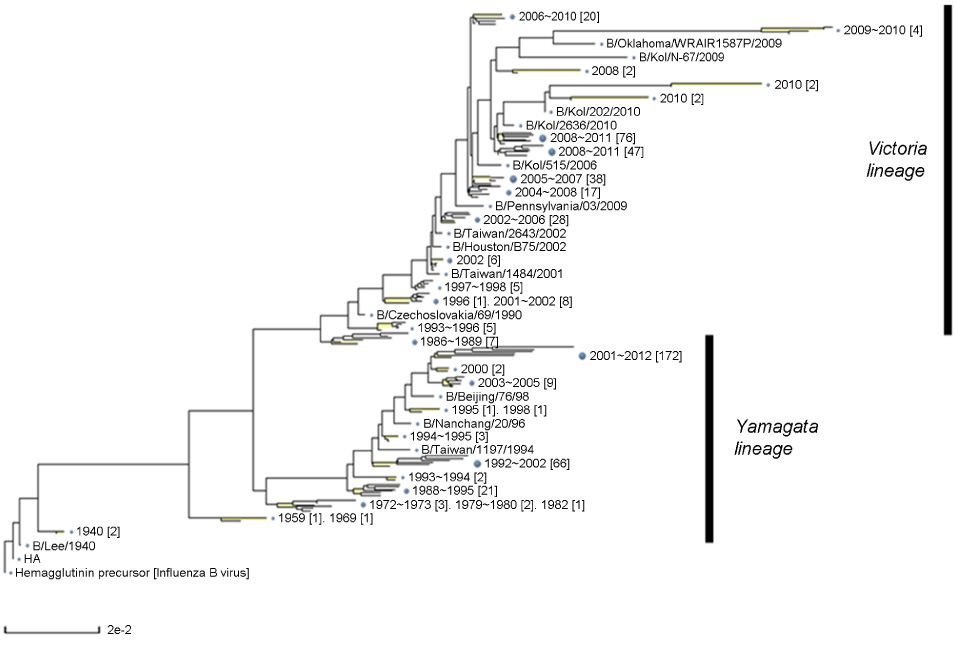
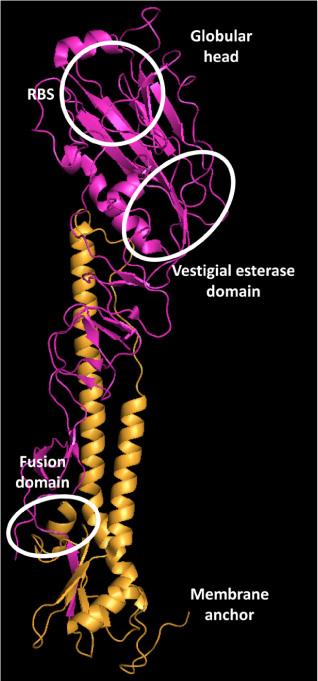
- Cross-reactive neutralizing antibodies against influenza A viruses have received attention for their potentials for prophylactic and therapeutic. These antibodies usually bind to relatively conserved stem domains of influenza hemagglutinin, one of surface glycoproteins responsible for viral binding to sialic acid-tagged cellular receptors and for membrane fusion to initiate a release process of viral genomes inside cells. Recently, a similar approach extended to influenza B viruses, which causing annual epidemics only in the human population, and some of human monoclonal antibodies exhibited promising efficacies against two antigenically diverged lineages of influenza B viruses. Moreover, one of these broadly neutralizing antibodies protected mice against both of influenza A and B challenges. Appropriate immunization may selectively enhance the efficacy of these antibodies, and this strategy may lead individuals to be prepared with broad immune responses against various influenza viruses.
MeSH Terms
-
Animals
Antibodies
Antibodies, Monoclonal
Antibodies, Neutralizing
Collodion
Genome, Viral
Hemagglutinins
Humans
Immunization
Influenza A virus
Influenza B virus
Influenza, Human
Membrane Fusion
Membrane Glycoproteins
Mice
Orthomyxoviridae
Vaccines
Antibodies
Antibodies, Monoclonal
Antibodies, Neutralizing
Collodion
Hemagglutinins
Membrane Glycoproteins
Vaccines
Figure
Cited by 3 articles
-
Age-Period-Cohort Analysis of Influenza in Koreans: the National Health Insurance Research Database, 2009–2018
Kyeong Hyang Byeon, Jaiyong Kim, Bo Youl Choi, Jin Yong Kim, Nakyoung Lee
J Korean Med Sci. 2020;35(18):e121. doi: 10.3346/jkms.2020.35.e121.Strategy for Developing Medical Arsenals by Modulation of Membrane Fusion Activity of Influenza Virus Hemagglutinin
Sangmoo Lee, Jin Il Kim, Ilseob Lee, Man-Seong Park
J Bacteriol Virol. 2013;43(4):337-341. doi: 10.4167/jbv.2013.43.4.337.Cell Culture-based Influenza Vaccines as Alternatives to Egg-based Vaccines
Ilseob Lee, Jin Il Kim, Man-Seong Park
J Bacteriol Virol. 2013;43(1):9-17. doi: 10.4167/jbv.2013.43.1.9.
Reference
-
1. Thompson WW, Shay DK, Weintraub E, Brammer L, Bridges CB, Cox NJ, et al. Influenza-associated hospitalizations in the United States. JAMA. 2004. 292:1333–1340.
Article2. Rota PA, Wallis TR, Harmon MW, Rota JS, Kendal AP, Nerome K. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology. 1990. 175:59–68.
Article3. Ambrose CS, Levin MJ. The rationale for quadrivalent influenza vaccines. Hum Vaccin Immunother. 2012. 8:81–88.
Article4. Ekiert DC, Wilson IA. Broadly neutralizing antibodies against influenza virus and prospects for universal therapies. Curr Opin Virol. 2012. 2:134–141.
Article5. Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, et al. Highly conserved protective epitopes on influenza B viruses. Science. 2012. 337:1343–1348.
Article6. Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009. 324:246–251.
Article7. Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science. 2011. 333:843–850.
Article8. Gubareva LV, Kaiser L, Hayden FG. Influenza virus neuraminidase inhibitors. Lancet. 2000. 355:827–835.
Article9. Lingwood D, McTamney PM, Yassine HM, Whittle JR, Guo X, Boyington JC, et al. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature. 2012. 489:566–570.
Article