Cancer Res Treat.
2011 Dec;43(4):205-211.
Systems Biology Approaches to Decoding the Genome of Liver Cancer
- Affiliations
-
- 1Department of Systems Biology, MD Anderson Cancer Center, The University of Texas, Houston, TX, USA. jlee@mdanderson.org
Abstract
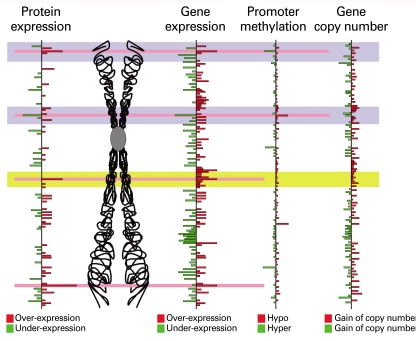
- Molecular classification of cancers has been significantly improved patient outcomes through the implementation of treatment protocols tailored to the abnormalities present in each patient's cancer cells. Breast cancer represents the poster child with marked improvements in outcome occurring due to the implementation of targeted therapies for estrogen receptor or human epidermal growth factor receptor-2 positive breast cancers. Important subtypes with characteristic molecular features as potential therapeutic targets are likely to exist for all tumor lineages including hepatocellular carcinoma (HCC) but have yet to be discovered and validated as targets. Because each tumor accumulates hundreds or thousands of genomic and epigenetic alterations of critical genes, it is challenging to identify and validate candidate tumor aberrations as therapeutic targets or biomarkers that predict prognosis or response to therapy. Therefore, there is an urgent need to devise new experimental and analytical strategies to overcome this problem. Systems biology approaches integrating multiple data sets and technologies analyzing patient tissues holds great promise for the identification of novel therapeutic targets and linked predictive biomarkers allowing implementation of personalized medicine for HCC patients.
Keyword
MeSH Terms
-
Biomarkers
Breast
Breast Neoplasms
Carcinoma, Hepatocellular
Child
Clinical Protocols
Epidermal Growth Factor
Epigenomics
Estrogens
Gene Expression Profiling
Genome
Genomics
Humans
Precision Medicine
Liver
Liver Neoplasms
Oligonucleotide Array Sequence Analysis
Prognosis
Proteomics
Systems Biology
Epidermal Growth Factor
Estrogens
Figure
Reference
-
1. Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005; 55:74–108. PMID: 15761078.
Article2. Bruix J, Boix L, Sala M, Llovet JM. Focus on hepatocellular carcinoma. Cancer Cell. 2004; 5:215–219. PMID: 15050913.
Article3. Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003; 362:1907–1917. PMID: 14667750.
Article4. Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002; 31:339–346. PMID: 12149612.
Article5. Lee JS, Thorgeirsson SS. Genome-scale profiling of gene expression in hepatocellular carcinoma: classification, survival prediction, and identification of therapeutic targets. Gastroenterology. 2004; 127(5 Suppl 1):S51–S55. PMID: 15508103.
Article6. Llovet JM, Bustamante J, Castells A, Vilana R, Ayuso Mdel C, Sala M, et al. Natural history of untreated nonsurgical hepatocellular carcinoma: rationale for the design and evaluation of therapeutic trials. Hepatology. 1999; 29:62–67. PMID: 9862851.
Article7. Calvet X, Bruix J, Ginés P, Bru C, Solé M, Vilana R, et al. Prognostic factors of hepatocellular carcinoma in the west: a multivariate analysis in 206 patients. Hepatology. 1990; 12(4 Pt 1):753–760. PMID: 2170267.
Article8. A new prognostic system for hepatocellular carcinoma: a retrospective study of 435 patients: the Cancer of the Liver Italian Program (CLIP) investigators. Hepatology. 1998; 28:751–755. PMID: 9731568.9. Kudo M, Chung H, Osaki Y. Prognostic staging system for hepatocellular carcinoma (CLIP score): its value and limitations, and a proposal for a new staging system, the Japan Integrated Staging Score (JIS score). J Gastroenterol. 2003; 38:207–215. PMID: 12673442.
Article10. Leung TW, Tang AM, Zee B, Lau WY, Lai PB, Leung KL, et al. Construction of the Chinese University Prognostic Index for hepatocellular carcinoma and comparison with the TNM staging system, the Okuda staging system, and the Cancer of the Liver Italian Program staging system: a study based on 926 patients. Cancer. 2002; 94:1760–1769. PMID: 11920539.11. Faratian D, Bown JL, Smith VA, Langdon SP, Harrison DJ. Cancer systems biology. Methods Mol Biol. 2010; 662:245–263. PMID: 20824475.
Article12. Kreeger PK, Lauffenburger DA. Cancer systems biology: a network modeling perspective. Carcinogenesis. 2010; 31:2–8. PMID: 19861649.
Article13. Sebolt-Leopold JS, English JM. Mechanisms of drug inhibition of signalling molecules. Nature. 2006; 441:457–462. PMID: 16724058.
Article14. Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008; 359:378–390. PMID: 18650514.
Article15. Cheng AL, Kang YK, Chen Z, Tsao CJ, Qin S, Kim JS, et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009; 10:25–34. PMID: 19095497.
Article16. Abou-Alfa GK, Johnson P, Knox JJ, Capanu M, Davidenko I, Lacava J, et al. Doxorubicin plus sorafenib vs doxorubicin alone in patients with advanced hepatocellular carcinoma: a randomized trial. JAMA. 2010; 304:2154–2160. PMID: 21081728.17. Nowell PC, Hungerford DA. Chromosome studies on normal and leukemic human leukocytes. J Natl Cancer Inst. 1960; 25:85–109. PMID: 14427847.18. Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992; 258:818–821. PMID: 1359641.
Article19. Moinzadeh P, Breuhahn K, Stützer H, Schirmacher P. Chromosome alterations in human hepatocellular carcinomas correlate with aetiology and histological grade: results of an explorative CGH meta-analysis. Br J Cancer. 2005; 92:935–941. PMID: 15756261.20. Kusano N, Shiraishi K, Kubo K, Oga A, Okita K, Sasaki K. Genetic aberrations detected by comparative genomic hybridization in hepatocellular carcinomas: their relationship to clinicopathological features. Hepatology. 1999; 29:1858–1862. PMID: 10347130.
Article21. Zimonjic DB, Keck CL, Thorgeirsson SS, Popescu NC. Novel recurrent genetic imbalances in human hepatocellular carcinoma cell lines identified by comparative genomic hybridization. Hepatology. 1999; 29:1208–1214. PMID: 10094966.
Article22. Poon TC, Wong N, Lai PB, Rattray M, Johnson PJ, Sung JJ. A tumor progression model for hepatocellular carcinoma: bioinformatic analysis of genomic data. Gastroenterology. 2006; 131:1262–1270. PMID: 17030195.
Article23. Pinkel D, Albertson DG. Comparative genomic hybridization. Annu Rev Genomics Hum Genet. 2005; 6:331–354. PMID: 16124865.
Article24. Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000; 403:503–511. PMID: 10676951.
Article25. Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, Hendrix M, et al. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature. 2000; 406:536–540. PMID: 10952317.
Article26. Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004; 40:667–676. PMID: 15349906.
Article27. Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002; 8:816–824. PMID: 12118244.
Article28. van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002; 415:530–536. PMID: 11823860.29. van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002; 347:1999–2009. PMID: 12490681.
Article30. Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998; 20:207–211. PMID: 9771718.
Article31. Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, et al. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet. 1999; 23:41–46. PMID: 10471496.
Article32. Sheehan KM, Calvert VS, Kay EW, Lu Y, Fishman D, Espina V, et al. Use of reverse phase protein microarrays and reference standard development for molecular network analysis of metastatic ovarian carcinoma. Mol Cell Proteomics. 2005; 4:346–355. PMID: 15671044.
Article33. Espina V, Mehta AI, Winters ME, Calvert V, Wulfkuhle J, Petricoin EF 3rd, et al. Protein microarrays: molecular profiling technologies for clinical specimens. Proteomics. 2003; 3:2091–2100. PMID: 14595807.
Article34. Haab BB. Antibody arrays in cancer research. Mol Cell Proteomics. 2005; 4:377–383. PMID: 15671041.
Article35. Liotta LA, Espina V, Mehta AI, Calvert V, Rosenblatt K, Geho D, et al. Protein microarrays: meeting analytical challenges for clinical applications. Cancer Cell. 2003; 3:317–325. PMID: 12726858.
Article36. Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet. 2011; 12:499–510. PMID: 21681211.
Article37. Hoshida Y, Villanueva A, Kobayashi M, Peix J, Chiang DY, Camargo A, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med. 2008; 359:1995–2004. PMID: 18923165.
Article38. Woo HG, Park ES, Cheon JH, Kim JH, Lee JS, Park BJ, et al. Gene expression-based recurrence prediction of hepatitis B virus-related human hepatocellular carcinoma. Clin Cancer Res. 2008; 14:2056–2064. PMID: 18381945.
Article39. Ye QH, Qin LX, Forgues M, He P, Kim JW, Peng AC, et al. Predicting hepatitis B virus-positive metastatic hepatocellular carcinomas using gene expression profiling and supervised machine learning. Nat Med. 2003; 9:416–423. PMID: 12640447.
Article40. Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006; 12:410–416. PMID: 16532004.
Article41. Van Dyke T, Jacks T. Cancer modeling in the modern era: progress and challenges. Cell. 2002; 108:135–144. PMID: 11832204.42. Klausner RD. Studying cancer in the mouse. Oncogene. 1999; 18:5249–5252. PMID: 10498876.
Article43. Lee JS, Chu IS, Mikaelyan A, Calvisi DF, Heo J, Reddy JK, et al. Application of comparative functional genomics to identify best-fit mouse models to study human cancer. Nat Genet. 2004; 36:1306–1311. PMID: 15565109.
Article44. Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu Q, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci U S A. 2010; 107:1437–1442. PMID: 20080689.
Article45. Lee KP, Lee JH, Kim TS, Kim TH, Park HD, Byun JS, et al. The Hippo-Salvador pathway restrains hepatic oval cell proliferation, liver size, and liver tumorigenesis. Proc Natl Acad Sci U S A. 2010; 107:8248–8253. PMID: 20404163.
Article46. Song H, Mak KK, Topol L, Yun K, Hu J, Garrett L, et al. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc Natl Acad Sci U S A. 2010; 107:1431–1436. PMID: 20080598.
Article47. Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009; 16:425–438. PMID: 19878874.48. Ferrando AA, Neuberg DS, Staunton J, Loh ML, Huard C, Raimondi SC, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002; 1:75–87. PMID: 12086890.
Article49. Eferl R, Ricci R, Kenner L, Zenz R, David JP, Rath M, et al. Liver tumor development. c-Jun antagonizes the proapoptotic activity of p53. Cell. 2003; 112:181–192. PMID: 12553907.50. Hilberg F, Aguzzi A, Howells N, Wagner EF. c-jun is essential for normal mouse development and hepatogenesis. Nature. 1993; 365:179–181. PMID: 8371760.
Article51. Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004; 20:781–810. PMID: 15473860.
Article52. Lee JS, Thorgeirsson SS. Functional and genomic implications of global gene expression profiles in cell lines from human hepatocellular cancer. Hepatology. 2002; 35:1134–1143. PMID: 11981763.
Article53. Lee JS, Grisham JW, Thorgeirsson SS. Comparative functional genomics for identifying models of human cancer. Carcinogenesis. 2005; 26:1013–1020. PMID: 15677630.
Article54. Lee JS, Thorgeirsson SS. Genetic profiling of human hepatocellular carcinoma. Semin Liver Dis. 2005; 25:125–132. PMID: 15918141.
Article55. Thorgeirsson SS, Lee JS, Grisham JW. Functional genomics of hepatocellular carcinoma. Hepatology. 2006; 43(2 Suppl 1):S145–S150. PMID: 16447291.
Article56. Thorgeirsson SS, Lee JS, Grisham JW. Molecular prognostication of liver cancer: end of the beginning. J Hepatol. 2006; 44:798–805. PMID: 16488507.
Article57. Thorgeirsson SS. Genomic decoding of hepatocellular carcinoma. Gastroenterology. 2006; 131:1344–1346. PMID: 17030203.
Article58. Paris PL, Andaya A, Fridlyand J, Jain AN, Weinberg V, Kowbel D, et al. Whole genome scanning identifies genotypes associated with recurrence and metastasis in prostate tumors. Hum Mol Genet. 2004; 13:1303–1313. PMID: 15138198.
Article59. Chin K, de Solorzano CO, Knowles D, Jones A, Chou W, Rodriguez EG, et al. In situ analyses of genome instability in breast cancer. Nat Genet. 2004; 36:984–988. PMID: 15300252.
Article60. Weiss MM, Kuipers EJ, Postma C, Snijders AM, Pinkel D, Meuwissen SG, et al. Genomic alterations in primary gastric adenocarcinomas correlate with clinicopathological characteristics and survival. Cell Oncol. 2004; 26:307–317. PMID: 15623941.
Article61. Carrasco DR, Tonon G, Huang Y, Zhang Y, Sinha R, Feng B, et al. High-resolution genomic profiles define distinct clinico-pathogenetic subgroups of multiple myeloma patients. Cancer Cell. 2006; 9:313–325. PMID: 16616336.
Article62. Rubio-Moscardo F, Climent J, Siebert R, Piris MA, Martin-Subero JI, Nieländer I, et al. Mantle-cell lymphoma genotypes identified with CGH to BAC microarrays define a leukemic subgroup of disease and predict patient outcome. Blood. 2005; 105:4445–4454. PMID: 15718413.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Genomic Profiling of Liver Cancer
- Systems biology approaches in asthma pharmacogenomics study
- Systems Biology: A Multi-Omics Integration Approach to Metabolism and the Microbiome
- Advances in Systems Biology Approaches for Autoimmune Diseases
- Precision medicine in nonalcoholic fatty liver disease: New therapeutic insights from genetics and systems biology