Healthc Inform Res.
2016 Apr;22(2):129-141. 10.4258/hir.2016.22.2.129.
Development of an Integrated Biospecimen Database among the Regional Biobanks in Korea
- Affiliations
-
- 1Department of Medical Informatics, Kyungpook National University, Daegu, Korea.
- 2Faculty of Medical Industry Convergence, Daegu Haany University, Gyeongsan, Korea. pulala@dhu.ac.kr
- KMID: 2166953
- DOI: http://doi.org/10.4258/hir.2016.22.2.129
Abstract
OBJECTIVES
This study developed an integrated database for 15 regional biobanks that provides large quantities of high-quality bio-data to researchers to be used for the prevention of disease, for the development of personalized medicines, and in genetics studies.
METHODS
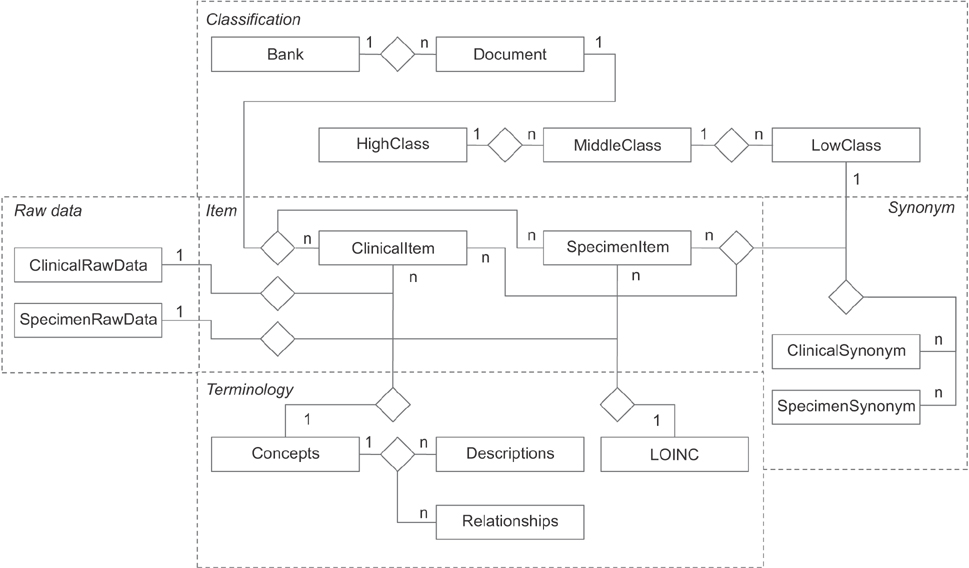
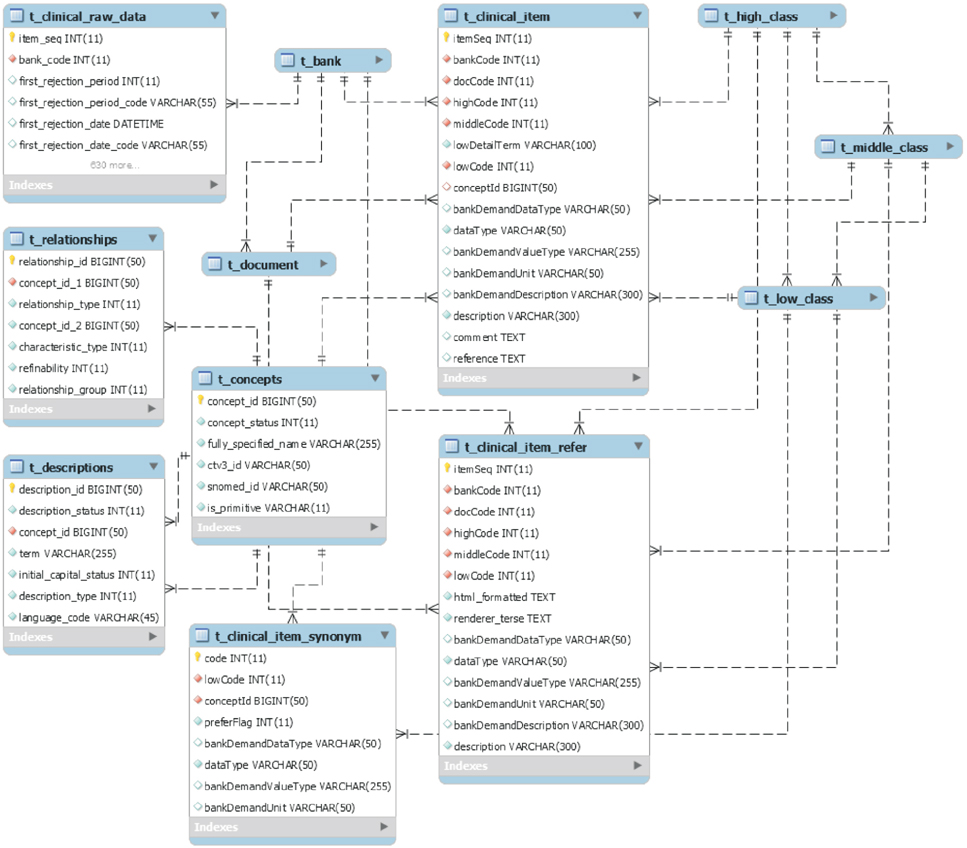
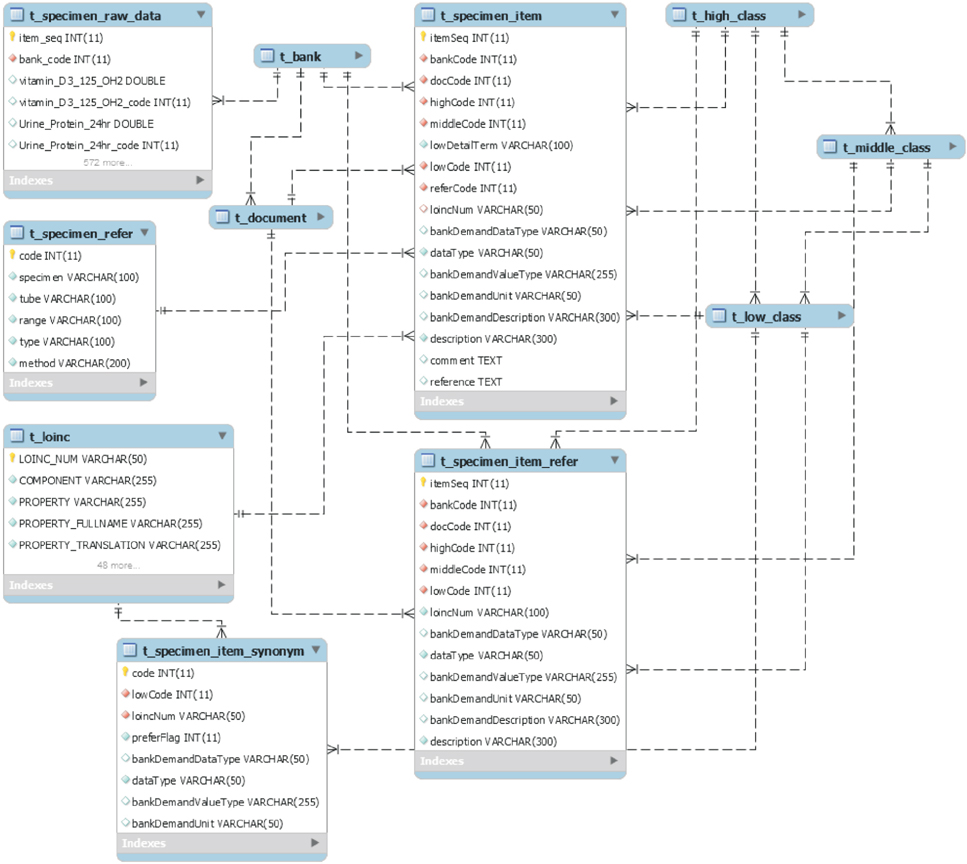
We collected raw data, managed independently by 15 regional biobanks, for database modeling and analyzed and defined the metadata of the items. We also built a three-step (high, middle, and low) classification system for classifying the item concepts based on the metadata. To generate clear meanings of the items, clinical items were defined using the Systematized Nomenclature of Medicine Clinical Terms, and specimen items were defined using the Logical Observation Identifiers Names and Codes. To optimize database performance, we set up a multi-column index based on the classification system and the international standard code.
RESULTS
As a result of subdividing 7,197,252 raw data items collected, we refined the metadata into 1,796 clinical items and 1,792 specimen items. The classification system consists of 15 high, 163 middle, and 3,588 low class items. International standard codes were linked to 69.9% of the clinical items and 71.7% of the specimen items. The database consists of 18 tables based on a table from MySQL Server 5.6. As a result of the performance evaluation, the multi-column index shortened query time by as much as nine times.
CONCLUSIONS
The database developed was based on an international standard terminology system, providing an infrastructure that can integrate the 7,197,252 raw data items managed by the 15 regional biobanks. In particular, it resolved the inevitable interoperability issues in the exchange of information among the biobanks, and provided a solution to the synonym problem, which arises when the same concept is expressed in a variety of ways.
MeSH Terms
Figure
Reference
-
1. Riegman PH, Morente MM, Betsou F, de Blasio P, Geary P. Marble Arch International Working Group on Biobanking for Biomedical Research. Biobanking for better healthcare. Mol Oncol. 2008; 2(3):213–222.
Article2. Jayasinghe SR, Mishra A, Van Daal A, Kwan E. Genetics and cardiovascular disease: design and development of a DNA biobank. Exp Clin Cardiol. 2009; 14(3):33–37.3. Founti P, Topouzis F, van Koolwijk L, Traverso CE, Pfeiffer N, Viswanathan AC. Biobanks and the importance of detailed phenotyping: a case study: the European Glaucoma Society Glauco GENE project. Br J Ophthalmol. 2009; 93(5):577–581.
Article4. Voidonikolas G, Gingras MC, Hodges S, McGuire AL, Chen C, Gibbs RA, et al. Developing a tissue resource to characterize the genome of pancreatic cancer. World J Surg. 2009; 33(4):723–731.
Article5. Olund G, Lindqvist P, Litton JE. BIMS: an information management system for biobanking in the 21st century. IBM Syst J. 2007; 46(1):171–182.
Article6. Saltz J, Oster S, Hastings S, Langella S, Kurc T, Sanchez W, et al. caGrid: design and implementation of the core architecture of the cancer biomedical informatics grid. Bioinformatics. 2006; 22(15):1910–1916.
Article7. Hirtzlin I, Dubreuil C, Preaubert N, Duchier J, Jansen B, Simon J, et al. An empirical survey on biobanking of human genetic material and data in six EU countries. Eur J Hum Genet. 2003; 11(6):475–488.
Article8. Yuille M, van Ommen GJ, Brechot C, Cambon-Thomsen A, Dagher G, Landegren U, et al. Biobanking for Europe. Brief Bioinform. 2008; 9(1):14–24.
Article9. Zika E, Gurwitz D, Ibarreta D. Pharmacogenetics and pharmacogenomics: state-of-the-art and potential socio-economic impacts in the EU. Brussels, Belgium: Joint Research Center, European Commission;2006.10. Austin MA, Harding S, McElroy C. Genebanks: a comparison of eight proposed international genetic databases. Community Genet. 2003; 6(1):37–45.
Article11. Spath MB, Grimson J. Applying the archetype approach to the database of a biobank information management system. Int J Med Inform. 2011; 80(3):205–226.
Article12. Watson PH, Wilson-McManus JE, Barnes RO, Giesz SC, Png A, Hegele RG, et al. Evolutionary concepts in biobanking - the BC BioLibrary. J Transl Med. 2009; 7:95.
Article13. Mora O, Bisbal J. BIMS: biomedical information management system [Internet]. [place unknown]: [publisher unknown];2013. cited at 2016 Mar 30. Available from: http://arxiv.org/abs/1303.5874.14. Muilu J, Peltonen L, Litton JE. The federated database: a basis for biobank-based post-genome studies, integrating phenome and genome data from 600,000 twin pairs in Europe. Eur J Hum Genet. 2007; 15(7):718–723.
Article15. Mohanty SK, Mistry AT, Amin W, Parwani AV, Pople AK, Schmandt L, et al. The development and deployment of Common Data Elements for tissue banks for translational research in cancer: an emerging standard based approach for the Mesothelioma Virtual Tissue Bank. BMC Cancer. 2008; 8:91.16. Cho SY, Hong EJ, Nam JM, Han B, Chu C, Park O. Opening of the national biobank of Korea as the infrastructure of future biomedical science in Korea. Osong Public Health Res Perspect. 2012; 3(3):177–184.
Article17. Park O, Cho SY, Shin SY, Park JS, Kim JW, Han BG. A strategic plan for the second phase (2013-2015) of the Korea biobank project. Osong Public Health Res Perspect. 2013; 4(2):107–116.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Opening of the National Biobank of Korea as the Infrastructure of Future Biomedical Science in Korea
- A Strategic Plan for the Second Phase (2013–2015) of the Korea Biobank Project
- Current Status, Challenges, Policies, and Bioethics of Biobanks
- ManBIF: a Program for Mining and Managing Biobank Impact Factor Data
- Management and Ethics of Biobank; Biorepository