Cancer Res Treat.
2004 Aug;36(4):207-213.
Microarray Applications in Cancer Research
- Affiliations
-
- 1Korean Hereditary Tumor Registry, Cancer Research Institute and Cancer Research Center, Seoul National University, Seoul, Korea. park@nccre.kr
- 2Research Institute and Hospital, National Cancer Center, Goyang, Gyeonggi, Korea.
Abstract
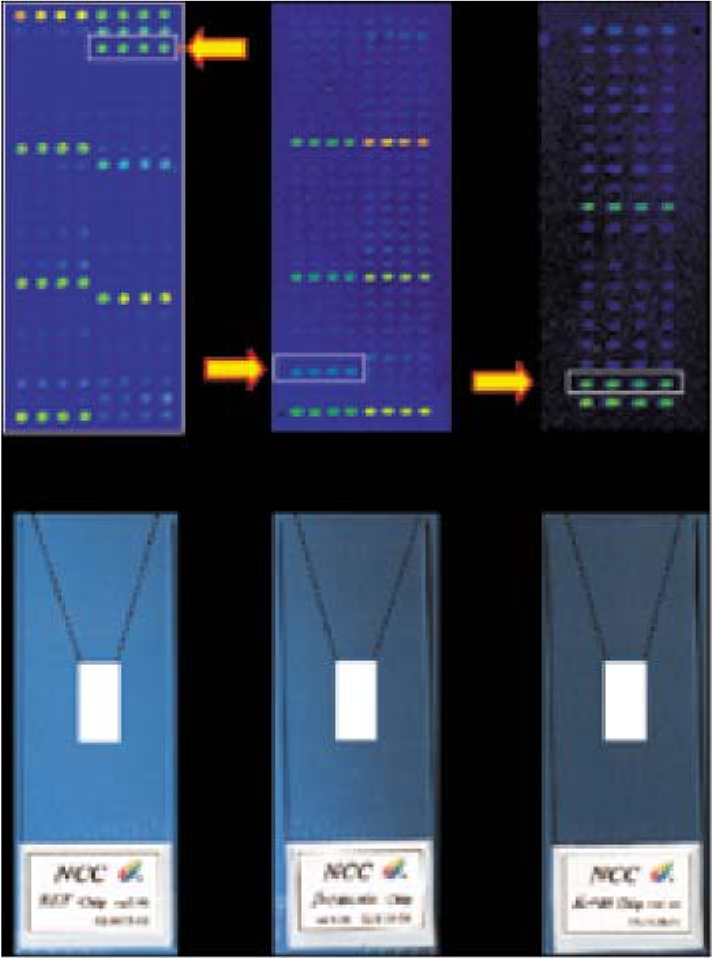
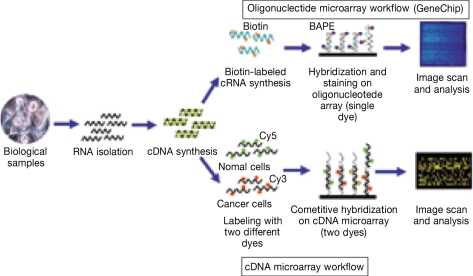
- DNA microarray technology permits simultaneous analysis of thousands of DNA sequences for genomic research and diagnostics applications. Microarray technology represents the most recent and exciting advance in the application of hybridization-based technology for biological sciences analysis. This review focuses on the classification (oligonucleotide vs. cDNA) and application (mutation-genotyping vs. gene expression) of microarrays. Oligonucleotide microarrays can be used both in mutation-genotyping and gene expression analysis, while cDNA microarrays can only be used in gene expression analysis. We review microarray mutation analysis, including examining the use of three oligonucleotide microarrays developed in our laboratory to determine mutations in RET, beta-catenin and K-ras genes. We also discuss the use of the Affymetrix GeneChip in mutation analysis. We review microarray gene expression analysis, including the classifying of such studies into four categories: class comparison, class prediction, class discovery and identification of biomarkers.
MeSH Terms
Figure
Reference
-
1. Schena M, Davis RW. Schena M, editor. Technology standards for microarray research. Microarray biochip technology. 2000. Massachusetts: Eaton publishing;p. 1–18.2. Russo G, Zegar C, Giordano A. Advantages and limitations of microarray technology in human cancer. Oncogene. 2003; 22:6497–6507. PMID: 14528274.
Article3. Yershov G, Barsky V, Belgovskiy A, Kirillov E, Kreindlin E, Ivanov I, Parinov S, Guschin D, Drobishev A, Dubiley S, Mirzabekov A. DNA analysis and diagnostics on oligonucleotide microchips. Proc Natl Acad Sci USA. 1996; 93:4913–4918. PMID: 8643503.
Article4. Wikman FP, Lu ML, Thykjaer T, Olesen SH, Andersen LD, Cordon-Cardo C, Orntoft TF. Evaluation of the performance of a p53 sequencing microarray chip using 140 previously sequenced bladder tumor samples. Clin Chem. 2000; 46:1555–1561. PMID: 11017932.
Article5. Hacia JG, Sun B, Hunt N, Edgemon K, Mosbrook D, Robbins C, Fodor SP, Tagle DA, Collins FS. Strategies for mutational analysis of the large multiexon ATM gene using high-density oligonucleotide arrays. Genome Res. 1998; 8:1245–1258. PMID: 9872980.
Article6. Kim IJ, Kang HC, Park JH, Shin Y, Ku JL, Yoo BC, Park JG. Hardiman G, editor. Development and application of an oligonucleotide microarray for mutational analysis. Microarrays methods and applications. 2003. Pennsylvania: DNA press;p. 250–271.7. Wen WH, Bernstein L, Lescallett J, Beazer-Barclay Y, Sullivan-Halley J, White M, Press MF. Comparison of TP53 mutations identified by oligonucleotide microarray and conventional DNA sequence analysis. Cancer Res. 2000; 60:2716–2722. PMID: 10825146.8. Kim IJ, Kang HC, Park JH, Ku JL, Lee JS, Kwon HJ, Yoon KA, Heo SC, Yang HY, Cho BY, Kim SY, Oh SK, Youn YK, Park DJ, Lee MS, Lee KW, Park JG. RET oligonucleotide microarray for the detection of RET mutations in multiple endocrine neoplasia type 2 syndromes. Clin Cancer Res. 2002; 8:457–463. PMID: 11839664.9. Lopez-Crapez E, Livache T, Marchand J, Grenier J. K-ras mutation detection by hybridization to a polypyrrole DNA chip. Clin Chem. 2001; 47:186–194. PMID: 11159765.
Article10. Gitan RS, Shi H, Chen CM, Yan PS, Huang TH. Methylation-specificoligonucleotide microarray: a new potential for high-throughput methylation analysis. Genome Res. 2002; 12:158–164. PMID: 11779841.11. Abraham SC, Nobukawa B, Giardiello FM, Hamilton SR, Wu TT. Sporadic fundic gland polyps: common gastric polyps arising through activating mutations in the beta-catenin gene. Am J Pathol. 2001; 158:1005–1010. PMID: 11238048.12. Abraham SC, Montgomery EA, Giardiello FM, Wu TT. Frequent beta-catenin mutations in juvenile nasopharyngeal angiofibromas. Am J Pathol. 2001; 158:1073–1078. PMID: 11238055.13. Saegusa M, Okayasu I. Frequent nuclear beta-catenin accumulation and associated mutations in endometrioid-type endometrial and ovarian carcinomas with squamous differentiation. J Pathol. 2001; 194:59–67. PMID: 11329142.14. Kim IJ, Kang HC, Park JH, Shin Y, Ku JL, Lim SB, Park SY, Jung SY, Kim HK, Park JG. Development and applications of a beta-catenin oligonucleotide microarray: beta-catenin mutations are dominantly found in the proximal colon cancers with microsatellite instability. Clin Cancer Res. 2003; 9:2920–2925. PMID: 12912937.15. Samowitz WS, Curtin K, Schaffer D, Robertson M, Leppert M, Slattery ML. Relationship of Ki-ras mutations in colon cancers to tumor location, stage, and survival: a population-based study. Cancer Epidemiol Biomarkers Prev. 2000; 9:1193–1197. PMID: 11097226.16. Andreyev HJ, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, et al. Kirsten ras mutations in patients with colorectal cancer: the 'RASCAL II' study. Br J Cancer. 2001; 85:692–696. PMID: 11531254.17. Macgregor PF, Squire JA. Application of microarrays to the analysis of gene expression in cancer. Clin Chem. 2002; 48:1170–1177. PMID: 12142369.
Article18. Warrington JA, Dee S, Trulson M. Schena M, editor. Large-scale genomic analysis using affymetrix GeneChip probe arrays. Microarray Biochip Technology. 2000. Massachusetts: Eaton Publishing;p. 201–220.19. Chou WH, Yan FX, Robbins-Weilert DK, Ryder TB, Liu WW, Perbost C, Fairchild M, de Leon J, Koch WH, Wedlund PJ. Comparison of two CYP2D6 genotyping methods and assessment of genotype-phenotype relationships. Clin Chem. 2003; 49:542–551. PMID: 12651805.
Article20. Khan J, Saal LH, Bittner ML, Chen Y, Trent JM, Meltzer PS. Expression profiling in cancer using cDNA microarrays. Electrophoresis. 1999; 20:223–229. PMID: 10197427.
Article21. Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, Brown EL. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996; 14:1675–1680. PMID: 9634850.
Article22. Schmidt U, Begley CG. Cancer diagnosis and microarrays. Int J Biochem Cell Biol. 2003; 35:119–124. PMID: 12479861.
Article23. Jain KK. Applications of biochips: from diagnostics to personalized medicine. Curr Opin Drug Discov Devel. 2004; 7:285–289.24. Hughes TR, Mao M, Jones AR, Burchard J, Marton MJ, Shannon KW, Lefkowitz SM, Ziman M, Schelter JM, Meyer MR, Kobayashi S, Davis C, Dai H, He YD, Stephaniants SB, Cavet G, Walker WL, West A, Coffey E, Shoemaker DD, Stoughton R, Blanchard AP, Friend SH, Linsley PS. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat Biotechnol. 2001; 19:342–347. PMID: 11283592.
Article25. Schulze A, Downward J. Navigating gene expression using microarrays--a technology review. Nat Cell Biol. 2001; 3:E190–E195. PMID: 11483980.26. Kurian KM, Watson CJ, Wyllie AH. DNA chip technology. J Pathol. 1999; 187:267–271. PMID: 10398077.27. Glanzer JG, Haydon PG, Eberwine JH. Expression profile analysis of neurodegenerative disease: advances in specificity and resolution. Neurochem Res. 2004; 29:1161–1168. PMID: 15176473.
Article28. Sotiriou C, Powles TJ, Dowsett M, Jazaeri AA, Feldman AL, Assersohn L, Gadisetti C, Libutti SK, Liu ET. Gene expression profiles derived from fine needle aspiration correlate with response to systemic chemotherapy in breast cancer. Breast Cancer Res. 2002; 4:R3. PMID: 12052255.
Article29. Park JH, Kim IJ, Kang HC, Shin Y, Park HW, Jang SG, Ku JL, Lim SB, Jeong SY, Park JG. Oligonucleotide microarray-based mutation detection of the K-ras gene in colorectal cancers using Competitive DNA Hybridization (CDH). Clin Chem. 2004; 50:1688–1691. PMID: 15331511.30. Wang DG, Fan JB, Siao CJ, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, Kruglyak L, Stein L, Hsie L, Topaloglou T, Hubbell E, Robinson E, Mittmann M, Morris MS, Shen N, Kilburn D, Rioux J, Nusbaum C, Rozen S, Hudson TJ, Lander ES, et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998; 280:1077–1082. PMID: 9582121.
Article31. Mei R, Galipeau PC, Prass C, Berno A, Ghandour G, Patil N, Wolff RK, Chee MS, Reid BJ, Lockhart DJ. Genome-wide detection of allelic imbalance using human SNPs and high-density DNA arrays. Genome Res. 2000; 10:1126–1137. PMID: 10958631.
Article32. Hacia JG, Brody LC, Chee MS, Fodor SP, Collins FS. Detection of heterozygous mutations in BRCA1 using high density oligonucleotide arrays and two-colour fluorescence analysis. Nat Genet. 1996; 14:441–447. PMID: 8944024.
Article33. Favis R, Day JP, Gerry NP, Phelan C, Narod S, Barany F. Universal DNA array detection of small insertions and deletions in BRCA1 and BRCA2. Nat Biotechnol. 2000; 18:561–564. PMID: 10802632.34. Ahrendt SA, Halachmi S, Chow JT, Wu L, Halachmi N, Yang SC, Wehage S, Jen J, Sidransky D. Rapid p53 sequence analysis in primary lung cancer using an oligonucleotide probe array. Proc Natl Acad Sci USA. 1999; 96:7382–7387. PMID: 10377423.
Article35. Simon R. Diagnostic and prognostic prediction using gene expression profiles in high-dimensional microarray data. Br J Cancer. 2003; 89:1599–1604. PMID: 14583755.
Article36. Simon R, Radmacher MD, Dobbin K, McShane LM. Pitfalls in the use of DNA microarray data for diagnostic and prognostic classification. J Natl Cancer Inst. 2003; 95:14–18. PMID: 12509396.
Article37. Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999; 286:531–537. PMID: 10521349.
Article38. Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M, Kallioniemi OP, Wilfond B, Borg A, Trent J. Gene-expression profiles in hereditary breast cancer. N Engl J Med. 2001; 344:539–548. PMID: 11207349.
Article39. Yuan Z, Sotsky Kent T, Weber TK. Differential expression of DOC-1 in microsatellite-unstable human colorectal cancer. Oncogene. 2003; 22:6304–6310. PMID: 13679870.
Article40. Lossos IS, Czerwinski DK, Alizadeh AA, Wechser MA, Tibshirani R, Botstein D, Levy R. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N Engl J Med. 2004; 350:1828–1837. PMID: 15115829.
Article41. Bvan de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictorof survival in breast cancer. N Engl J Med. 2002; 347:1999–2009. PMID: 12490681.42. Huang E, Cheng SH, Dressman H, Pittman J, Tsou MH, Horng CF, Bild A, Iversen ES, Liao M, Chen CM, West M, Nevins JR, Huang AT. Gene expression predictors of breast cancer outcomes. Lancet. 2003; 361:1590–1596. PMID: 12747878.
Article43. Kihara C, Tsunoda T, Tanaka T, Yamana H, Furukawa Y, Ono K, Kitahara O, Zembutsu H, Yanagawa R, Hirata K, Takagi T, Nakamura Y. Prediction of sensitivity of esophageal tumors to adjuvant chemotherapy by cDNA microarray analysis of gene-expression profiles. Cancer Res. 2001; 61:6474–6479. PMID: 11522643.44. Mariadason JM, Arango D, Shi Q, Wilson AJ, Corner GA, Nicholas C, Aranes MJ, Lesser M, Schwartz EL, Augenlicht LH. Gene expression profiling-based prediction of response of colon carcinoma cells to 5-fluorouracil and camptothecin. Cancer Res. 2003; 63:8791–8812. PMID: 14695196.45. Kim HK, Choi IJ, Kim HS, Kim JH, Kim E, Park IS, Chun JH, Kim IH, Kim IJ, Kang HC, Park JH, Bae JM, Lee JS, Park JG. DNA microarray analysis of the correlation between gene expression patterns and acquired resistance to 5-FU/cisplatin in gastric cancer. Biochem Biophys Res Commun. 2004; 316:781–789. PMID: 15033468.
Article46. Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J Jr, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000; 403:503–511. PMID: 10676951.
Article47. Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, Hendrix M, Radmacher M, Simon R, Yakhini Z, Ben-Dor A, Sampas N, Dougherty E, Wang E, Marincola F, Gooden C, Lueders J, Glatfelter A, Pollock P, Carpten J, Gillanders E, Leja D, Dietrich K, Beaudry C, Berens M, Alberts D, Sondak V. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature. 2000; 406:536–540. PMID: 10952317.
Article48. Kang HC, Kim IJ, Park JH, Shin Y, Ku JL, Jung MS, Yoo BC, Kim HK, Park JG. Identification of genes with differential expression in acquired drug-resistant gastric cancer cells using high-density oligonucleotide microarrays. Clin Cancer Res. 2004; 10:272–284. PMID: 14734480.
Article49. Chun JH, Kim HK, Kim E, Kim IH, Kim JH, Chang HJ, Choi IJ, Lim HS, Kim IJ, Kang HC, Park JH, Bae JM, Park JG. Increased expression of metallothionein is associated with irinotecan resistance in gastric cancer. Cancer Res. 2004; 64:4703–4706. PMID: 15256434.
Article50. Wreesmann VB, Sieczka EM, Socci ND, Hezel M, Belbin TJ, Childs G, Patel SG, Patel KN, Tallini G, Prystowsky M, Shaha AR, Kraus D, Shah JP, Rao PH, Ghossein R, Singh B. Genome-wide profiling of papillary thyroid cancer identifies MUC1 as an independent prognostic marker. Cancer Res. 2004; 64:3780–3789. PMID: 15172984.
Article51. Koopmann J, Buckhaults P, Brown DA, Zahurak ML, Sato N, Fukushima N, Sokoll LJ, Chan DW, Yeo CJ, Hruban RH, Breit SN, Kinzler KW, Vogelstein B, Goggins M. Serum macrophage inhibitory cytokine 1 as a marker of pancreatic and other periampullary cancers. Clin Cancer Res. 2004; 10:2386–2392. PMID: 15073115.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Role of Microarray in Cancer Diagnosis
- A DNA Microarray LIMS System for Integral Genomic Analysis of Multi-Platform Microarrays
- Predicting 5-Year Survival Status of Patients with Breast Cancer based on Supervised Wavelet Method
- Microarray Data Analysis of Perturbed Pathways in Breast Cancer Tissues
- Significant Gene Selection Using Integrated Microarray Data Set with Batch Effect