Korean J Phys Anthropol.
2016 Mar;29(1):27-34. 10.11637/kjpa.2016.29.1.27.
Effect of MITF-M and MITF-A Overexpression on the Dendrtic Formation in Melanocytes
- Affiliations
-
- 1Department of Anatomy, College of Medicine, Chungnam National University, Daejeon, Korea. yhlee@cnu.ac.kr
- 2Department of Dermatology, College of Medicine, Chungnam National University, Daejeon, Korea.
- KMID: 2161138
- DOI: http://doi.org/10.11637/kjpa.2016.29.1.27
Abstract
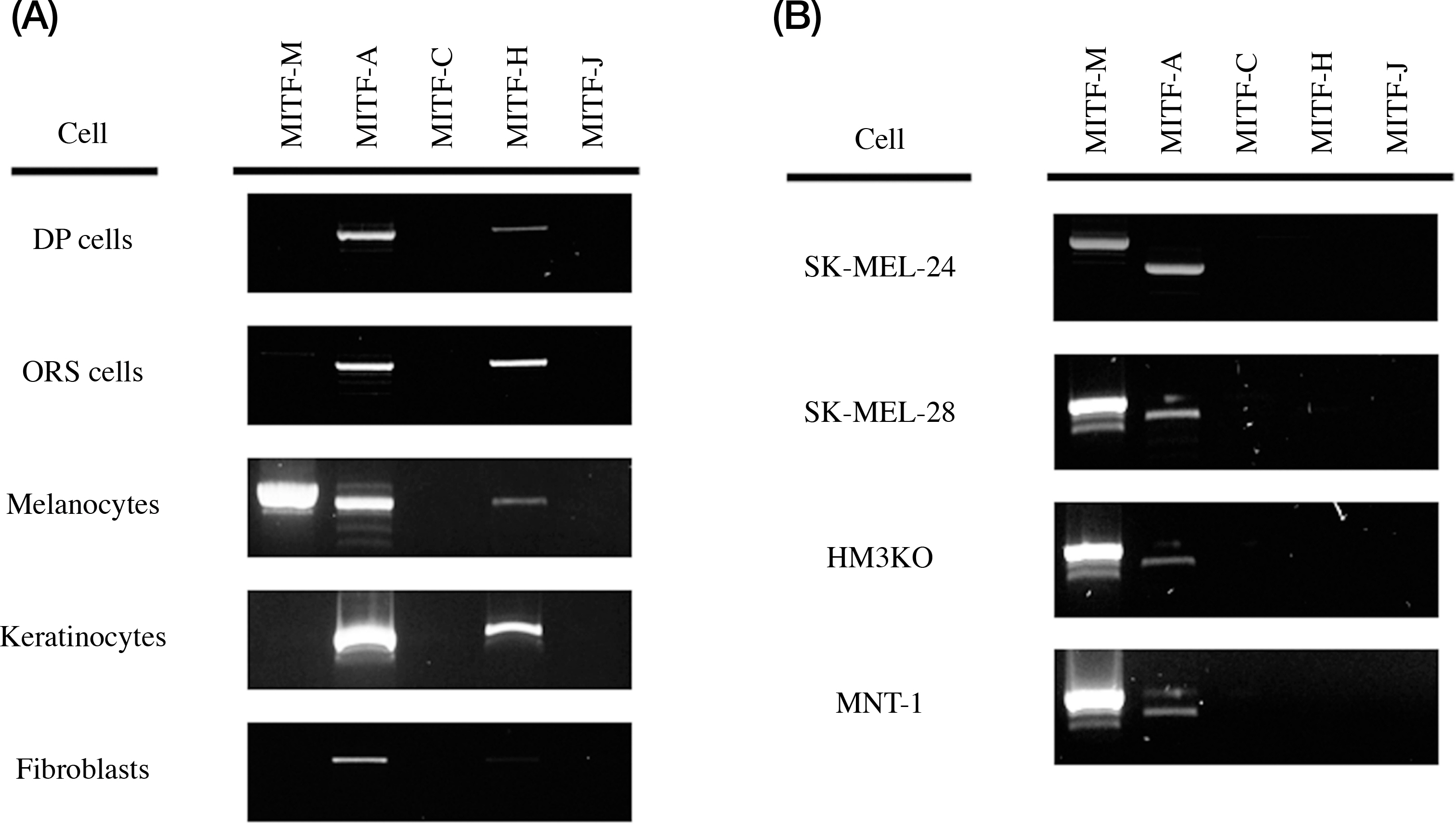
- The microphthalmia-associated transcription factor (MITF), has been described as the master regulator of the basic helix-loop-helix leucine zipper family, involves melanogenesis in melanocytes. MITF consists of at least six isoforms, called MITF-M, MITF-A, MITF-B, MITF-C, MITF-H, and MITF-J. Previously, we found that not only MITF-M is expressed in the human hair follicle, but also MITF-A, MITF-C, MITF-H, and MITF-J isoforms are expressed in the skin. The aim of this study was to conform the MITF isoforms expressed in human skin, and investigate novel role of MITF isoforms in the melanocytes. Expression of MITF-M and MITF-A was found in primary melanoctyes and the melanoma cell lines. Interestingly, when MITF-M and MITF-A were overexpressed in the SK-MEL-24 melanoma cells by adenoviral transfection, length of the dendrites, serves as the principal conduit for melanosomes transfer, was significantly increased in the MITF-M overexpressed cells compared with the control group, and number of the dendtrites was significantly increased in the MITF-A overexpressed cells. A signal molecule involve in actin polymerization during dendrite formation, Rac1, was increased in the SK-MEL-24 melanoma cells treated with adenoviral MITF-M and MITF-A vectors. These results suggest that MITF-M and MITF-A induce dendrite formation via Rac1 signaling in the melanocytes.
Keyword
MeSH Terms
Figure
Reference
-
References
1. Hodgkinson CA, Moore KJ, Nakayama A, Steingrimsson A, Copeland NG, Jenkins NA, et al. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993; 74:395–404.
Article2. Widlund HR, Fisher DE. Microphthalamia-associated transcription factor: a critical regulator of pigment cell development and survival. Oncogene. 2003; 22:3035–41.
Article3. Steingrimsson E, Copeland NG, Jenkins NA. Melanocytes and the microphthalmia transcription factor network. Annu Rev Genet. 2004; 38:365–411.4. Price ER, Horstmann MA, Wells AG, Weilbaecher KN, Takemoto CM, Landis MW, et al. Alpha-melanocyte-stimulating hormone signaling regulates expression of microphthalmia, a gene deficient in Waardenburg syndrome. J Biol Chem. 1988; 273:33042–7.5. Bertolotto C, Abbe P, Timothy J, Bille HK, Fisher DE, Ortonne JP, et al. Microphthalmia gene product as a signal transducer in cAMP-induced differentiation of melanocytes. J Cell Biol. 1998; 142:827–35.
Article6. Goding CR. Mitf from neural crest to melanoma: signal transduction and transcription in the melanocyte lineage. Genes Dev. 2000; 14:1712–28.
Article7. Hershey CL, Fisher DE. Genomic analysis of the Microphthalmia locus and identification of the MITF-J/Mitf-J isoform. Gene. 2005; 347:73–82.
Article8. Tachibana M. Cochlear Melanocytes and MITF Signaling. J Invest Dermatol Symp Proc. 2001; 6:95–8.
Article9. Vachtenheim J, Novotna AH. Expression of genes for microphthalmia isoforms, Pax3 and MSG1, in human melanomas. Cell Mol Biol. 1999; 45:1075–82.10. Ooishi R, Shirai M, Funaba M, Murakami M. Microphthalmia-associated transcription factor is required for mature myotube formation. Biochim Biophys Acta. 2012; 1820:76–83.
Article11. Lee SH, Lee JH, Lee JH, Kim DK. Involvement of MITF-A, an alternative isoform of mi transcription factor, on the expression of tryptase gene in human mast cells. Exp Mol Med. 2010; 42:366–75.12. Johnson SL, Nguyen AN, Lister JA. mitfa is required at multiple stages of melanocyte differentiation but not to establish the melanocyte stem cell. Dev Biol. 2011; 350:405–13.
Article13. Dorsky RI, Moon RT, Raible DW. Environmental signals and cell fate specification in premigratory neural crest. Bioassay. 2000; 22:708–16.
Article14. Jimbow K, Quevedo WC, Prota G, Fitzpatrick TB. Biology of melanocytes. In: Freedberg IM, Eisen AZ, Wolff K, Aus-ten KF, Goldsmith LA, Katz SI. Dermatology in general medicine, 5th ed. .,. New York: McGraw-Hill;1999.15. Cardinali G, Ceccarelli S, Kovacs D, Aspite N, Lotti LV, Torrisi MR, et al. Keratinocyte growth factor promotes melanosome transfer to keratinocytes. J Invest Dermatol. 2005; 25:1190–9.
Article16. Gilchrest BA, Park HY, Eller M, Yaar M. The photobiology of the tanning response. Nordlund JJ, Boissy RE, Hearing VJ, King RA, Ortonne JP, editors. editors.The pigmentary system physiology and pathophysiology. New York: Oxford University Press;1998.17. Costin GE, Hearing VJ. Human skin pigmentation: melanocytes modulate skin color in response to stress. FASEB J. 2007; 21:976–94.
Article18. Ridley AJ, Hall A. The small GTP-binding protein Rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992; 70:389–99.
Article19. Ridley AJ, Hall A. Signal transduction pathways regulating Rhomedicated stress fiber formation: requirement for a tyrosine kinase. EMBO J. 1994; 13:2600–10.20. Ridley AJ, Paterson HF, Johnston CL. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992; 70:401–10.
Article21. Imokawa G. Autocrine and paracrine regulation of melanocytes in human skin and in pigmentary disorders. Pigment Cell Res. 2004; 17:96–110.
Article22. Herlyn M, Mancianti ML, Jambrosic J, Bolen JB, Koprow-ski H. Regulatory factors that determine growth and phenotype of normal human melanocytes. Exp Cell Res. 1988; 179:322–31.23. Masaru M, Yasuhiro I, Masayuki F. Expression and transcriptional activity of alternative splice variants of Mitf exon 6. Mol Cell Biochem. 2007; 303:251–57.24. Scott G. Rac and rho: the story behind melanocyte dendrite formation. Pigment Cell Res. 2002; 15:322–30.
Article25. Sander EE, Klooster JP, Delft S, Kammen RA, Collard JG. Rac downregulates Rho Activity: Reciprocal Balance between both GTPases determines cellular morphology and migratory behavior. J Cell Biol. 1999; 147:1009–21.26. Jalink K, Corven EJ, Hengeveld T, Morii N, Narumiya S, Moolenaar WH. Inhibition of lysophosphatidate and thrombin-induced neurite retraction and neuronal cell rounding by ADP ribosylation of the small GTPbinding protein Rho. J Cell Biol. 1994; 126:801–10.
Article27. Katoh H, Aoki J, Ichikawa A, Negishi M. RhoA-binding Kinase ROKa induces neurite retraction. J Biol Chem. 1998; 273:2489–92.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Involvement of MITF-A, an alternative isoform of mi transcription factor, on the expression of tryptase gene in human mast cells
- Recombinant AAV Vector with MITF-M Promoter for Melanoma Gene Therapy
- Modulation of Melanogenesis by Heme Oxygenase-1 via p53 in Normal Human Melanocytes
- BMP4-Induced Differentiation of Human Hair Follicle Neural Crest Stem Cells into Precursor Melanocytes from Hair Follicle Bulge
- Changes in Expressions of Endothelin-1, Microphthalmia Associated Transcription Factor, and Protease-activated Receptor-2 after Ultraviolet Irradiation