Korean J Leg Med.
2016 Feb;40(1):27-31. 10.7580/kjlm.2016.40.1.27.
Mixture Patterned Short Tandem Repeat Profile in a Perimortem Transfused Patient
- Affiliations
-
- 1Department of Forensic Medicine, Seoul National University College of Medicine, Seoul, Korea. sdlee@snu.ac.kr
- 2Institute of Forensic Science, Seoul National University College of Medicine, Seoul, Korea.
- 3Medical Examiner's Office, National Forensic Service, Wonju, Korea.
- KMID: 2155728
- DOI: http://doi.org/10.7580/kjlm.2016.40.1.27
Abstract
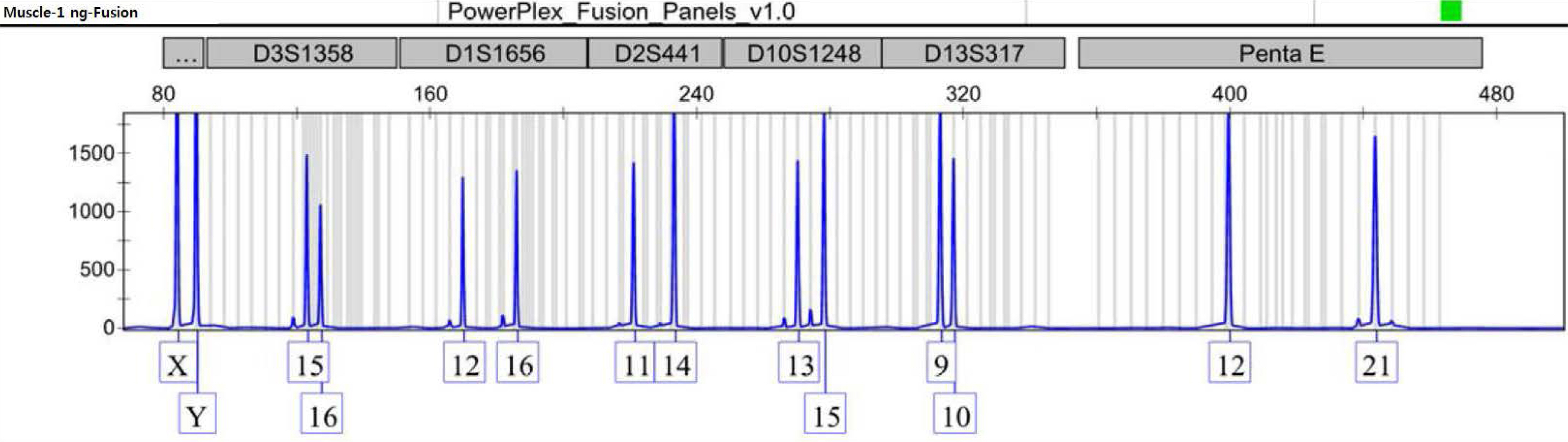
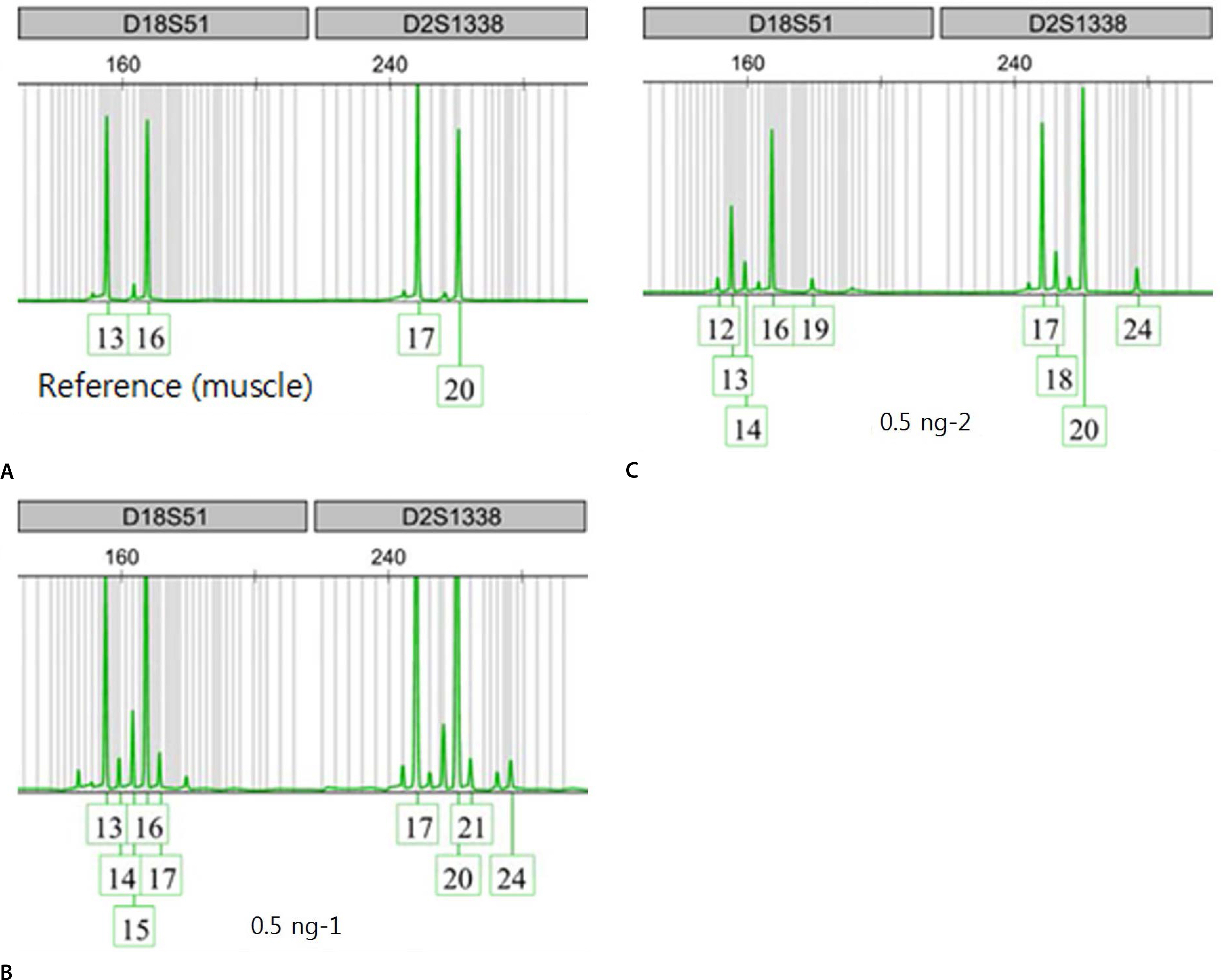
- Recently, it has been reported that transfused patients can generate admixture-like genetic profiles. As genetic material of the donor can survive for a reasonable time after transfusion, the recipient's genomic DNA is likely to have a mixture pattern. An autopsy case of a man transfused perimortem generated a mixture patterned short tandem repeat profile. Notably, the patient was transfused mostly with nuclear-deficient cells, limiting the donor genetic material available for the recipient. As a result, mixture-like patterns were observed consistently, regardless of change in input DNA content; the sample DNA content, which was serially diluted, ranged from 1 ng to 0.0625 ng. The distributions of foreign peaks appeared to be irreproducible, showing stochastic behaviors throughout the genotyped results. This study suggests that a cautious approach is required when genotyping of a patient who has undergone recent transfusion. One must consider the possibility of obtaining a mixture patterned profile in such patients, and therefore, choose parenchymal organs or tissues for reliable results.
MeSH Terms
Figure
Reference
-
1.Wenk RE., Chiafari PA. DNA typing of recipient blood after massive transfusion. Transfusion. 1997. 37:1108–10.
Article2.Graham EA., Tsokos M., Rutty GN. Can post-mortem blood be used for DNA profiling after peri-mortem blood transfusion? Int J Legal Med. 2007. 121:18–23.
Article3.Dauber EM., Dorner G., Mitterbauer M, et al. Discrepant results of samples taken from different tissues of a single individual. Int Congr Ser. 2004. 1261:48–9.
Article4.Vietor HE., Hallensleben E., van Bree SP, et al. Survival of donor cells 25 years after intrauterine transfusion. Blood. 2000. 95:2709–14.5.Gong MN., Sai Y., Zhou W, et al. Genotyping patients with recent blood transfusions. Epidemiology. 2003. 14:744–7.
Article6.Lee TH., Montalvo L., Chrebtow V, et al. Quantitation of genomic DNA in plasma and serum samples: higher concentrations of genomic DNA found in serum than in plasma. Transfusion. 2001. 41:276–82.
Article7.Schechter GP., Whang-Peng J., McFarland W. Circulation of donor lymphocytes after blood transfusion in man. Blood. 1977. 49:651–6.
Article8.Utter GH., Reed WF., Lee TH, et al. Transfusion-associated microchimerism. Vox Sang. 2007. 93:188–95.
Article9.Jung JY., Park SH., Park SW, et al. Mixed DNA profiles in transfused deceased case. Korean J Sci Crim Invest. 2015. 9:206–12.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Rapid Prenatal Detection of Down and Edwards Syndromes by Fluorescent Polymerase Chain Reaction with Short Tandem Repeat Markers
- Sequence Generation and Genotyping of 15 Autosomal STR Markers Using Next Generation Sequencing
- DNA Profiling via Short Tandem Repeat Analysis by Using Serum Samples
- Instability at Short Tandem Repeats in Lymphoblastoid Cell Lines
- Comparison of the Effects of Patterned and Conventional Panretinal Photocoagulation on Diabetic Retinopathy