Clin Exp Vaccine Res.
2016 Jan;5(1):41-49. 10.7774/cevr.2016.5.1.41.
In silico analysis of an envelope domain III-based multivalent fusion protein as a potential dengue vaccine candidate
- Affiliations
-
- 1Department of Molecular and Cellular Sciences, Faculty of Advanced Sciences and Technology, Pharmaceutical Sciences Branch, Islamic Azad University (IAUPS), Tehran, Iran.
- 2Department of Molecular Genetics, Faculty of Biological Sciences, Tarbiat Modares University, Tehran, Iran. sadeghma@modares.ac.ir
- 3Blood Transfusion Research Center, High Institute for Research and Education in Transfusion Medicine, Tehran, Iran.
- KMID: 2152696
- DOI: http://doi.org/10.7774/cevr.2016.5.1.41
Abstract
- PURPOSE
Dengue virus infection is now a global problem. Currently, there is no licensed vaccine or proven antiviral treatment against this virus. All four serotypes (1-4) of dengue virus can infect human. An effective dengue vaccine should be tetravalent to induce protective immune responses against all four serotypes. Most of dengue vaccine candidates are monovalent, or in the form of physically mixed multivalent formulations. Recently envelope protein domain III of virus is considered as a vaccine candidate, which plays critical roles in the most important viral activities. Development of a tetravalent protein subunit vaccine is very important for equal induction of immune system and prevention of unbalanced immunity. Here, we have presented and used a rational approach to design a tetravalent dengue vaccine candidate.
MATERIALS AND METHODS
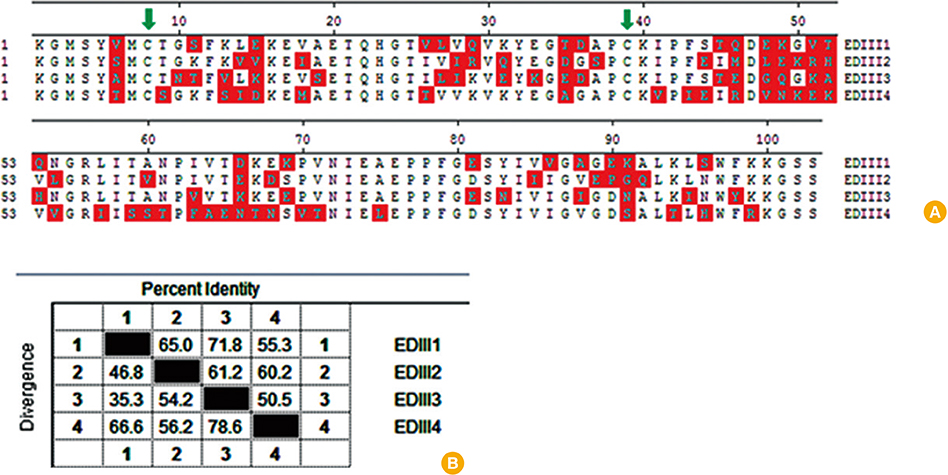
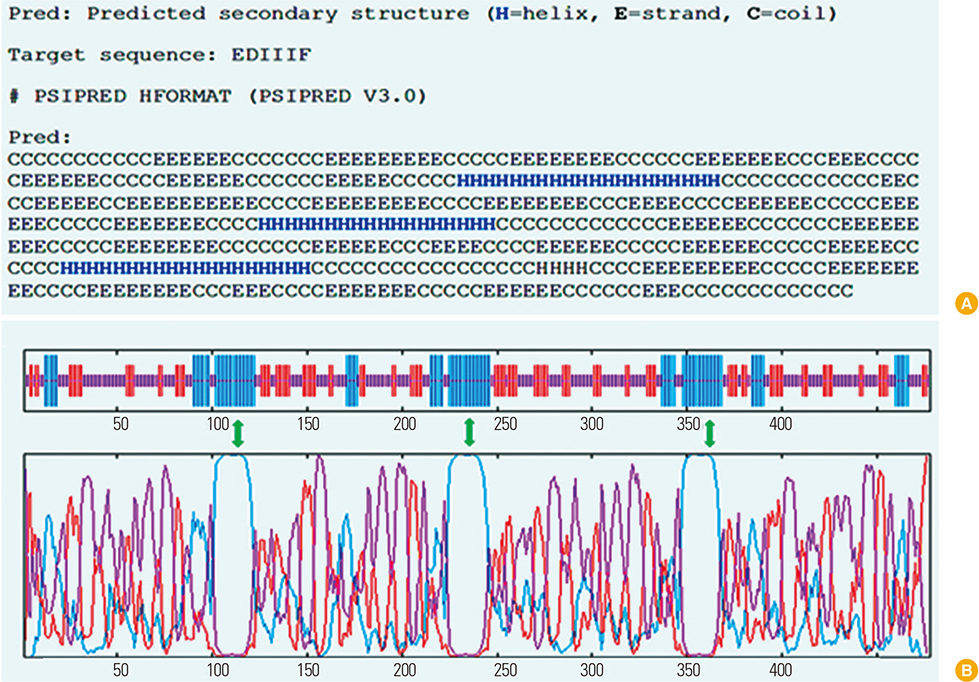
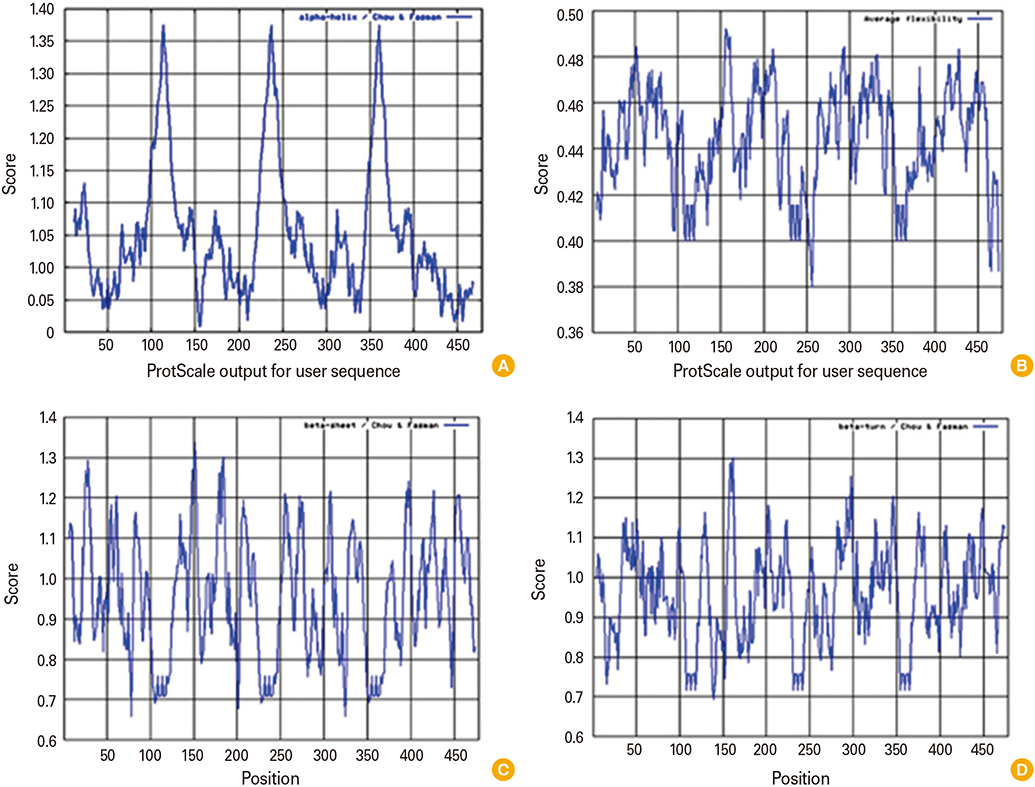
We designed a multi domain antigen by fusing four consensus domain III sequences together with appropriate hydrophobic linkers and used several types of bioinformatics software and neural networks to predict structural and immunological properties of the designed tetravalent antigen.
RESULTS
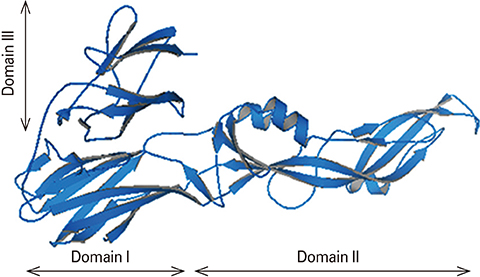
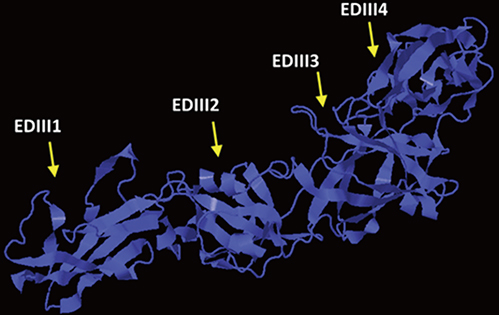
We designed a tetravalent protein (EDIIIF) based on domain III of dengue virus envelope protein. According to the results of the bioinformatics analysis, the constructed models for EDIIIF protein were structurally stable and potentially immunogenic.
CONCLUSION
The designed tetravalent protein can be considered as a potential dengue vaccine candidate. The presented approach can be used for rational design and in silico evaluation of chimeric dengue vaccine candidates.
Keyword
MeSH Terms
Figure
Reference
-
1. Kuno G, Chang GJ, Tsuchiya KR, Karabatsos N, Cropp CB. Phylogeny of the genus Flavivirus. J Virol. 1998; 72:73–83.2. Halstead SB. Dengue. Lancet. 2007; 370:1644–1652.
Article3. Rothman AL. Dengue: defining protective versus pathologic immunity. J Clin Invest. 2004; 113:946–951.
Article4. Fields BN, Knipe DM, Howley PM. Fields' virology. 5th ed. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins;2007.5. Crill WD, Chang GJ. Localization and characterization of flavivirus envelope glycoprotein cross-reactive epitopes. J Virol. 2004; 78:13975–13986.
Article6. Roehrig JT, Bolin RA, Kelly RG. Monoclonal antibody mapping of the envelope glycoprotein of the dengue 2 virus, Jamaica. Virology. 1998; 246:317–328.
Article7. Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004; 427:313–319.
Article8. Crill WD, Roehrig JT. Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to Vero cells. J Virol. 2001; 75:7769–7773.
Article9. Chavez JH, Silva JR, Amarilla AA, Moraes Figueiredo LT. Domain III peptides from flavivirus envelope protein are useful antigens for serologic diagnosis and targets for immunization. Biologicals. 2010; 38:613–618.
Article10. Swaminathan S, Batra G, Khanna N. Dengue vaccines: state of the art. Expert Opin Ther Pat. 2010; 20:819–835.
Article11. Puigbo P, Guzman E, Romeu A, Garcia-Vallve S. OPTIMIZER: a web server for optimizing the codon usage of DNA sequences. Nucleic Acids Res. 2007; 35:W126–W131.
Article12. Vincze T, Posfai J, Roberts RJ. NEBcutter: a program to cleave DNA with restriction enzymes. Nucleic Acids Res. 2003; 31:3688–3691.
Article13. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415.
Article14. Garnier J, Gibrat JF, Robson B. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 1996; 266:540–553.15. Arai R, Ueda H, Kitayama A, Kamiya N, Nagamune T. Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein Eng. 2001; 14:529–532.
Article16. Amani J, Mousavi SL, Rafati S, Salmanian AH. In silico analysis of chimeric espA, eae and tir fragments of Escherichia coli O157:H7 for oral immunogenic applications. Theor Biol Med Model. 2009; 6:28.
Article17. Koraka P, Martina BE, Osterhaus AD. Bioinformatics in new generation flavivirus vaccines. J Biomed Biotechnol. 2010; 2010:864029.
Article18. Holbrook MR, Shope RE, Barrett AD. Use of recombinant E protein domain III-based enzyme-linked immunosorbent assays for differentiation of tick-borne encephalitis serocomplex flaviviruses from mosquito-borne flaviviruses. J Clin Microbiol. 2004; 42:4101–4110.
Article19. Fahimi H, Allahyari H, Hassan ZM, Sadeghizadeh M. Dengue virus type-3 envelope protein domain III; expression and immunogenicity. Iran J Basic Med Sci. 2014; 17:836–843.20. Burgess-Brown NA, Sharma S, Sobott F, Loenarz C, Oppermann U, Gileadi O. Codon optimization can improve expression of human genes in Escherichia coli: a multi-gene study. Protein Expr Purif. 2008; 59:94–102.
Article21. Tong JC, Tan TW, Ranganathan S. Methods and protocols for prediction of immunogenic epitopes. Brief Bioinform. 2007; 8:96–108.
Article22. Kolaskar AS, Tongaonkar PC. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990; 276:172–174.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Prospects for dengue vaccines for travelers
- A potent multivalent vaccine for modulation of immune system in atherosclerosis: an in silico approach
- Development of a Novel Subunit Vaccine Targeting Fusobacterium nucleatum FomA Porin Based on In Silico Analysis
- Animal models for dengue vaccine development and testing
- Identifying immunodominant multi-epitopes from the envelope glycoprotein of the Lassa mammarenavirus as vaccine candidate for Lassa fever