Immune Netw.
2009 Dec;9(6):248-254. 10.4110/in.2009.9.6.248.
Tiul1 and TGIF are Involved in Downregulation of TGFbeta1-induced IgA Isotype Expression
- Affiliations
-
- 1Department of Molecular Bioscience, School of Bioscience and Biotechnology, Kangwon National University, Chuncheon 200-701, Korea. phkim@kangwon.ac.kr
- KMID: 2150653
- DOI: http://doi.org/10.4110/in.2009.9.6.248
Abstract
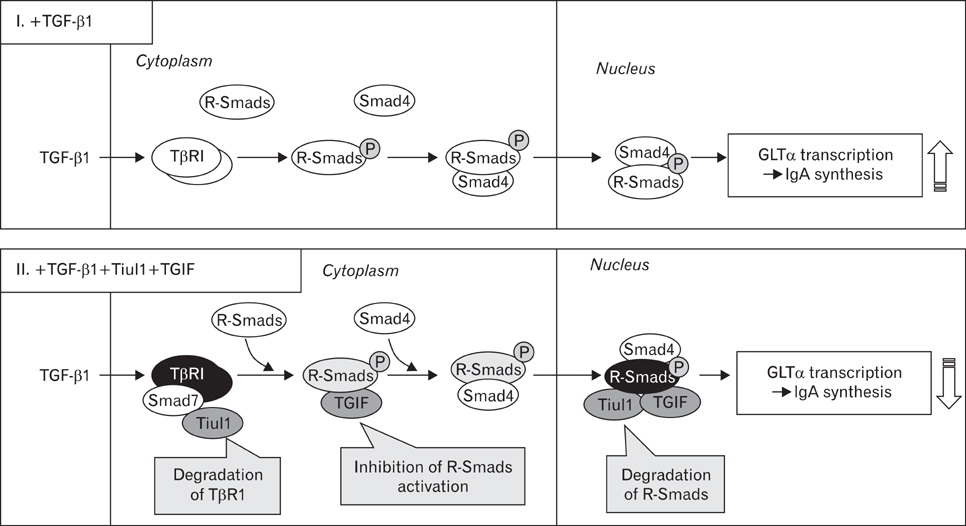
- TGF-beta1 is well known to induce Ig germ-line alpha (GLalpha) transcription and subsequent IgA isotype class switching recombination (CSR). Homeodomain protein TG-interacting factor (TGIF) and E3-ubiquitin ligases TGIF interacting ubiquitin ligase 1 (Tiul1) are implicated in the negative regulation of TGF-beta signaling. In the present study, we investigated the roles of Tiul1 and TGIF in TGFbeta1-induced IgA CSR. We found that over-expression of Tiul1 decreased TGFbeta1-induced GLalpha promoter activity and strengthened the inhibitory effect of Smad7 on the promoter activity. Likewise, overexpression of TGIF also diminished GLalpha promoter activity and further strengthened the inhibitory effect of Tiul1, suggesting that Tiul1 and TGIF can down-regulate TGFbeta1-induced GLalpha expression. In parallel, overexpression of Tiul1 decreased the expression of endogenous IgA CSR-predicitive transcripts (GLT(alpha), PST(alpha), and CT(alpha)) and TGFbeta1-induced IgA secretion, but not GLT(gamma3) and IgG3 secretion. Here, over-expressed TGIF further strengthened the inhibitory effect of Tiul1. These results suggest that Tiul1 and TGIF act as negatively regulators in TGFbeta1-induced IgA isotype expression.
MeSH Terms
Figure
Cited by 1 articles
-
SUMO Proteins are not Involved in TGF-β1-induced, Smad3/4-mediated Germline α Transcription, but PIASy Suppresses it in CH12F3-2A B Cells
Sang-Hoon Lee, Pyeung-Hyeun Kim, Sang-Muk Oh, Jung-Hwan Park, Yung-Choon Yoo, Junglim Lee, Seok-Rae Park
Immune Netw. 2014;14(6):321-327. doi: 10.4110/in.2014.14.6.321.
Reference
-
1. Lagna G, Hata A, Hemmati-Brivanlou A, Massagué J. Partnership between DPC4 and SMAD proteins in TGF-beta signalling pathways. Nature. 1996. 383:832–836.
Article2. Wu RY, Zhang Y, Feng XH, Derynck R. Heteromeric and homomeric interactions correlate with signaling activity and functional cooperativity of Smad3 and Smad4/DPC4. Mol Cell Biol. 1997. 17:2521–2528.
Article3. Zhang Y, Musci T, Derynck R. The tumor suppressor Smad4/DPC 4 as a central mediator of Smad function. Curr Biol. 1997. 7:270–276.
Article4. Nakao A, Imamura T, Souchelnytskyi S, Kawabata M, Ishisaki A, Oeda E, Tamaki K, Hanai J, Heldin CH, Miyazono K, ten Dijke P. TGF-beta receptor-mediated signalling through Smad2, Smad3 and Smad4. EMBO J. 1997. 16:5353–5362.
Article5. Hayashi H, Abdollah S, Qiu Y, Cai J, Xu YY, Grinnell BW, Richardson MA Jr, Topper JN, Gimbrone MA Jr, Wrana JL, Falb D. The MAD-related protein Smad7 associates with the TGFbeta receptor and functions as an antagonist of TGFbeta signaling. Cell. 1997. 89:1165–1173.
Article6. Imamura T, Takase M, Nishihara A, Oeda E, Hanai J, Kawabata M, Miyazono K. Smad6 inhibits signalling by the TGF-beta superfamily. Nature. 1997. 389:622–626.7. Nakao A, Afrakhte M, Morén A, Nakayama T, Christian JL, Heuchel R, Itoh S, Kawabata M, Heldin NE, Heldin CH, ten Dijke P. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature. 1997. 389:631–635.
Article8. Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, Miyazono K. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J Biol Chem. 2001. 276:12477–12480.
Article9. Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, Wrana JL. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell. 2000. 6:1365–1375.
Article10. Kim KI, Baek SH. SUMOylation code in cancer development and metastasis. Mol Cells. 2006. 22:247–253.11. Zhu H, Kavsak P, Abdollah S, Wrana JL, Thomsen GH. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature. 1999. 400:687–693.
Article12. Zhao M, Qiao M, Oyajobi BO, Mundy GR, Chen D. E3 ubiquitin ligase Smurf1 mediates core-binding factor alpha1/Runx2 degradation and plays a specific role in osteoblast differentiation. J Biol Chem. 2003. 278:27939–27944.
Article13. Jin YH, Jeon EJ, Li QL, Lee YH, Choi JK, Kim WJ, Lee KY, Bae SC. Transforming growth factor-beta stimulates p300-dependent RUNX3 acetylation, which inhibits ubiquitination-mediated degradation. J Biol Chem. 2004. 279:29409–29417.
Article14. Lin X, Liang M, Feng XH. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-beta signaling. J Biol Chem. 2000. 275:36818–36822.
Article15. Zhang Y, Chang C, Gehling DJ, Hemmati-Brivanlou A, Derynck R. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc Natl Acad Sci U S A. 2001. 98:974–979.
Article16. Koinuma D, Shinozaki M, Komuro A, Goto K, Saitoh M, Hanyu A, Ebina M, Nukiwa T, Miyazawa K, Imamura T, Miyazono K. Arkadia amplifies TGF-beta superfamily signalling through degradation of Smad7. EMBO J. 2003. 22:6458–6470.
Article17. Seo SR, Lallemand F, Ferrand N, Pessah M, L'hoste S, Camonis J, Atfi A. The novel E3 ubiquitin ligase Tiul1 associates with TGIF to target Smad2 for degradation. EMBO J. 2004. 23:3780–3792.
Article18. Wotton D, Lo RS, Lee S, Massague J. A Smad transcriptional corepressor. Cell. 1999. 97:29–39.
Article19. Wotton D, Lo RS, Swaby LA, Massagué J. Multiple modes of repression by the Smad transcriptional corepressor TGIF. J Biol Chem. 1999. 274:37105–37110.
Article20. Seo SR, Ferrand N, Faresse N, Prunier C, Abécassis L, Pessah M, Bourgeade MF, Atfi A. Nuclear retention of the tumor suppressor cPML by the homeodomain protein TGIF restricts TGF-beta signaling. Mol Cell. 2006. 23:547–559.
Article21. Park SR, Lee JH, Kim PH. Smad3 and Smad4 mediate transforming growth factor-beta1-induced IgA expression in murine B lymphocytes. Eur J Immunol. 2001. 31:1706–1715.
Article22. Park SR, Lee EK, Kim BC, Kim PH. p300 cooperates with Smad3/4 and Runx3 in TGFbeta1-induced IgA isotype expression. Eur J Immunol. 2003. 33:3386–3392.
Article23. Choi SH, Seo GY, Nam EH, Jeon SH, Kim HA, Park JB, Kim PH. Opposing effects of Arkadia and Smurf on TGFbeta1-induced IgA isotype expression. Mol Cells. 2007. 24:283–287.24. Li SC, Rothman PB, Zhang J, Chan C, Hirsh D, Alt FW. Expression of I mu-C gamma hybrid germline transcripts subsequent to immunoglobulin heavy chain class switching. Int Immunol. 1994. 6:491–497.
Article25. Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000. 102:553–563.
Article26. Kinoshita K, Harigai M, Fagarasan S, Muramatsu M, Honjo T. A hallmark of active class switch recombination: transcripts directed by I promoters on looped-out circular DNAs. Proc Natl Acad Sci U S A. 2001. 98:12620–12623.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- TGFbeta1 Effect on Survival of Anticancer Drug - resistant L1210 Sublines
- Further Characterization of Activin A-induced IgA Response in Murine B Lymphocytes
- Characterization of Mouse B Lymphoma Cells (CH12F3-2A) for the Study of IgA Isotype Switching
- Lactoferrin Combined with Retinoic Acid Stimulates B1 Cells to Express IgA Isotype and Gut-homing Molecules
- Preferential Expression of IgA Isotype Switching-associated Transcripts in Mouse Intestinal Lymphoid Tissues