Korean J Urol.
2015 Feb;56(2):109-116. 10.4111/kju.2015.56.2.109.
A clinicogenetic model to predict lymph node invasion by use of genome-based biomarkers from exome arrays in prostate cancer patients
- Affiliations
-
- 1Department of Urology, Seoul National University Bundang Hospital, Seongnam, Korea. ssbyun@snubh.org
- 2Department of Electrical and Computer Engineering, Seoul National University, Seoul, Korea.
- 3School of Electrical Engineering, Korea University, Seoul, Korea.
- 4Biomedical Research Institute, Seoul National University Bundang Hospital, Seongnam, Korea.
- KMID: 2133274
- DOI: http://doi.org/10.4111/kju.2015.56.2.109
Abstract
- PURPOSE
Genetic variations among prostate cancer (PCa) patients who underwent radical prostatectomy (RP) and pelvic lymph node dissection were evaluated to predict lymph node invasion (LNI). Exome arrays were used to develop a clinicogenetic model that combined clinical data related to PCa and individual genetic variations.
MATERIALS AND METHODS
We genotyped 242,186 single-nucleotide polymorphisms (SNPs) by using a custom HumanExome BeadChip v1.0 (Illumina Inc.) from the blood DNA of 341 patients with PCa. The genetic data were analyzed to calculate an odds ratio as an estimate of the relative risk of LNI. We compared the accuracies of the multivariate logistic model incorporating clinical factors between the included and excluded selected SNPs. The Cox proportional hazard models with or without genetic factors for predicting biochemical recurrence (BCR) were analyzed.
RESULTS
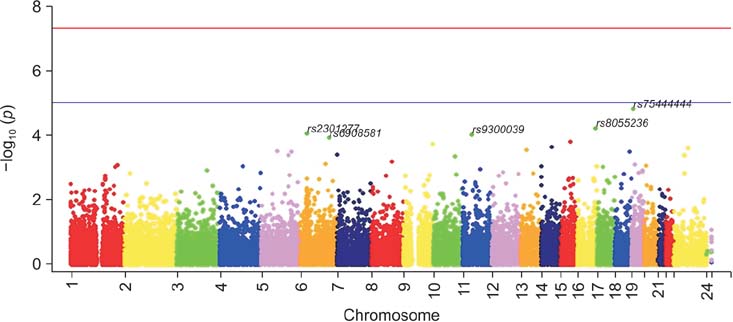
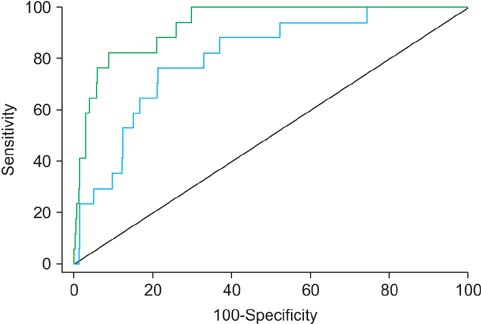
The genetic analysis indicated that five SNPs (rs75444444, rs8055236, rs2301277, rs9300039, and rs6908581) were significant for predicting LNI in patients with PCa. When a multivariate model incorporating clinical factors was devised to predict LNI, the predictive accuracy of the multivariate model was 80.7%. By adding genetic factors in the aforementioned multivariate model, the predictive accuracy increased to 93.2% (p=0.006). These genetic variations were significant factors for predicting BCR after adjustment for other variables and after adding the predictive gain to BCR.
CONCLUSIONS
Based on the results of the exome array, the selected SNPs were predictors for LNI. The addition of individualized genetic information effectively enhanced the predictive accuracy of LNI and BCR among patients with PCa who underwent RP.
MeSH Terms
-
Aged
Biomarkers, Tumor/*genetics
Biopsy
DNA, Neoplasm/genetics
Exome
Gene Frequency
Genome
Genotype
Humans
Lymph Node Excision
Lymph Nodes/pathology
Lymphatic Metastasis
Male
Middle Aged
*Models, Genetic
Neoplasm Invasiveness
Polymorphism, Single Nucleotide
Predictive Value of Tests
Prospective Studies
Prostatectomy
Prostatic Neoplasms/*genetics/pathology/surgery
Biomarkers, Tumor
DNA, Neoplasm
Figure
Reference
-
1. Pak S, Park S, Ryu J, Hong S, Song SH, You D, et al. Preoperative factors predictive of posterolateral extracapsular extension after radical prostatectomy. Korean J Urol. 2013; 54:824–829.2. Cooperberg MR, Moul JW, Carroll PR. The changing face of prostate cancer. J Clin Oncol. 2005; 23:8146–8151.3. Abdollah F, Suardi N, Gallina A, Bianchi M, Tutolo M, Passoni N, et al. Extended pelvic lymph node dissection in prostate cancer: a 20-year audit in a single center. Ann Oncol. 2013; 24:1459–1466.4. Abdollah F, Sun M, Thuret R, Budaus L, Jeldres C, Graefen M, et al. Decreasing rate and extent of lymph node staging in patients undergoing radical prostatectomy may undermine the rate of diagnosis of lymph node metastases in prostate cancer. Eur Urol. 2010; 58:882–892.5. Abdollah F, Karnes RJ, Suardi N, Cozzarini C, Gandaglia G, Fossati N, et al. Predicting survival of patients with node-positive prostate cancer following multimodal treatment. Eur Urol. 2014; 65:554–562.6. Walz J, Bladou F, Rousseau B, Laroche J, Salem N, Gravis G, et al. Head to head comparison of nomograms predicting probability of lymph node invasion of prostate cancer in patients undergoing extended pelvic lymph node dissection. Urology. 2012; 79:546–551.7. Briganti A, Larcher A, Abdollah F, Capitanio U, Gallina A, Suardi N, et al. Updated nomogram predicting lymph node invasion in patients with prostate cancer undergoing extended pelvic lymph node dissection: the essential importance of percentage of positive cores. Eur Urol. 2012; 61:480–487.8. Hansen J, Rink M, Bianchi M, Kluth LA, Tian Z, Ahyai SA, et al. External validation of the updated Briganti nomogram to predict lymph node invasion in prostate cancer patients undergoing extended lymph node dissection. Prostate. 2013; 73:211–218.9. Heidenreich A, Pfister D, Thuer D, Brehmer B. Percentage of positive biopsies predicts lymph node involvement in men with low-risk prostate cancer undergoing radical prostatectomy and extended pelvic lymphadenectomy. BJU Int. 2011; 107:220–225.10. Touijer K, Rabbani F, Otero JR, Secin FP, Eastham JA, Scardino PT, et al. Standard versus limited pelvic lymph node dissection for prostate cancer in patients with a predicted probability of nodal metastasis greater than 1%. J Urol. 2007; 178:120–124.11. Briganti A, Chun FK, Salonia A, Zanni G, Scattoni V, Valiquette L, et al. Validation of a nomogram predicting the probability of lymph node invasion among patients undergoing radical prostatectomy and an extended pelvic lymphadenectomy. Eur Urol. 2006; 49:1019–1026.12. Abdollah F, Schmitges J, Sun M, Tian Z, Briganti A, Shariat SF, et al. A critical assessment of the value of lymph node dissection at radical prostatectomy: A population-based study. Prostate. 2011; 71:1587–1594.13. Briganti A, Karakiewicz PI, Chun FK, Gallina A, Salonia A, Zanni G, et al. Percentage of positive biopsy cores can improve the ability to predict lymph node invasion in patients undergoing radical prostatectomy and extended pelvic lymph node dissection. Eur Urol. 2007; 51:1573–1581.14. Eeles RA, Olama AA, Benlloch S, Saunders EJ, Leongamornlert DA, Tymrakiewicz M, et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet. 2013; 45:385–391. 391e1–391e2.15. Huang SP, Huang LC, Ting WC, Chen LM, Chang TY, Lu TL, et al. Prognostic significance of prostate cancer susceptibility variants on prostate-specific antigen recurrence after radical prostatectomy. Cancer Epidemiol Biomarkers Prev. 2009; 18:3068–3074.16. McNeal JE, Redwine EA, Freiha FS, Stamey TA. Zonal distribution of prostatic adenocarcinoma. Correlation with histologic pattern and direction of spread. Am J Surg Pathol. 1988; 12:897–906.17. Cookson MS, Aus G, Burnett AL, Canby-Hagino ED, D'Amico AV, Dmochowski RR, et al. Variation in the definition of biochemical recurrence in patients treated for localized prostate cancer: the American Urological Association Prostate Guidelines for Localized Prostate Cancer Update Panel report and recommendations for a standard in the reporting of surgical outcomes. J Urol. 2007; 177:540–545.18. Hedrick PW. Gametic disequilibrium measures: proceed with caution. Genetics. 1987; 117:331–341.19. Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001; 68:978–989.20. Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004; 74:765–769.21. Donovan MJ, Hamann S, Clayton M, Khan FM, Sapir M, Bayer-Zubek V, et al. Systems pathology approach for the prediction of prostate cancer progression after radical prostatectomy. J Clin Oncol. 2008; 26:3923–3929.22. Svatek RS, Jeldres C, Karakiewicz PI, Suardi N, Walz J, Roehrborn CG, et al. Pre-treatment biomarker levels improve the accuracy of post-prostatectomy nomogram for prediction of biochemical recurrence. Prostate. 2009; 69:886–894.23. Witte JS. Prostate cancer genomics: towards a new understanding. Nat Rev Genet. 2009; 10:77–82.24. Williams KA, Terry KL, Tworoger SS, Vitonis AF, Titus LJ, Cramer DW. Polymorphisms of MUC16 (CA125) and MUC1 (CA15.3) in relation to ovarian cancer risk and survival. PLoS One. 2014; 9:e88334.25. Arlt A, Schafer H. Role of the immediate early response 3 (IER3) gene in cellular stress response, inflammation and tumorigenesis. Eur J Cell Biol. 2011; 90:545–552.26. Qiu Y, Morii E, Tomita Y, Zhang B, Matsumura A, Kitaichi M, et al. Prognostic significance of pre B cell leukemia transcription factor 2 (PBX2) expression in non-small cell lung carcinoma. Cancer Sci. 2009; 100:1198–1209.27. Heidenreich A, Bellmunt J, Bolla M, Joniau S, Mason M, Matveev V, et al. EAU guidelines on prostate cancer. Part 1: screening, diagnosis, and treatment of clinically localised disease. Eur Urol. 2011; 59:61–71.28. Briganti A, Blute ML, Eastham JH, Graefen M, Heidenreich A, Karnes JR, et al. Pelvic lymph node dissection in prostate cancer. Eur Urol. 2009; 55:1251–1265.29. van Houten ME, Gooren LJ. Differences in reproductive endocrinology between Asian men and Caucasian men: a literature review. Asian J Androl. 2000; 2:13–20.30. Song C, Ro JY, Lee MS, Hong SJ, Chung BH, Choi HY, et al. Prostate cancer in Korean men exhibits poor differentiation and is adversely related to prognosis after radical prostatectomy. Urology. 2006; 68:820–824.